Colorectal Cancer-Associated Immune Exhaustion Involves T and B Lymphocytes and Conventional NK Cells and Correlates With a Shorter Overall Survival
- PMID: 34975867
- PMCID: PMC8716410
- DOI: 10.3389/fimmu.2021.778329
Colorectal Cancer-Associated Immune Exhaustion Involves T and B Lymphocytes and Conventional NK Cells and Correlates With a Shorter Overall Survival
Abstract
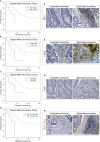
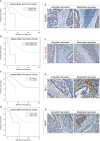
Colorectal cancer (CRC) is one of the most common cancer worldwide, with a growing impact on public health and clinical management. Immunotherapy has shown promise in the treatment of advanced cancers, but needs to be improved for CRC, since only a limited fraction of patients is eligible for treatment, and most of them develop resistance due to progressive immune exhaustion. Here, we identify the transcriptional, molecular, and cellular traits of the immune exhaustion associated with CRC and determine their relationships with the patient's clinic-pathological profile. Bioinformatic analyses of RNA-sequencing data of 594 CRCs from TCGA PanCancer collection, revealed that, in the wide range of immune exhaustion genes, those coding for PD-L1, LAG3 and T-bet were associated (Cramér's V=0.3) with MSI/dMMR tumors and with a shorter overall survival (log-rank test: p=0.0004, p=0.0014 and p=0.0043, respectively), whereas high levels of expression of EOMES, TRAF1, PD-L1, FCRL4, BTLA and SIGLEC6 were associated with a shorter overall survival (log-rank test: p=0.0003, p=0.0188, p=0.0004, p=0.0303, p=0.0052 and p=0.0033, respectively), independently from the molecular subtype of CRC. Expression levels of PD-L1, PD-1, LAG3, EOMES, T-bet, and TIGIT were significantly correlated with each other and associated with genes coding for CD4+ and CD8+CD3+ T cell markers and NKp46+CD94+EOMES+T-bet+ cell markers, (OR >1.5, p<0.05), which identify a subset of group 1 innate lymphoid cells, namely conventional (c)NK cells. Expression of TRAF1 and BTLA co-occurred with both T cell markers, CD3γ, CD3δ, CD3ε, CD4, and B cell markers, CD19, CD20 and CD79a (OR >2, p<0.05). Expression of TGFβ1 was associated only with CD4+ and CD8+CD3ε+ T cell markers (odds ratio >2, p<0.05). Expression of PD-L2 and IDO1 was associated (OR >1.5, p<0.05) only with cNK cell markers, whereas expression of FCRL4, SIGLEC2 and SIGLEC6 was associated (OR >2.5; p<0.05) with CD19+CD20+CD79a+ B cell markers. Morphometric examination of immunostained CRC tissue sections, obtained from a validation cohort of 53 CRC patients, substantiated the biostatistical findings, showing that the highest percentage of immune exhaustion gene expressing cells were found in tumors from short-term survivors and that functional exhaustion is not confined to T lymphocytes, but also involves B cells, and cNK cells. This concept was strengthened by CYBERSORTx analysis, which revealed the expression of additional immune exhaustion genes, in particular FOXP1, SIRT1, BATF, NR4A1 and TOX, by subpopulations of T, B and NK cells. This study provides novel insight into the immune exhaustion landscape of CRC and emphasizes the need for a customized multi-targeted therapeutic approach to overcome resistance to current immunotherapy.
Keywords: B cell exhaustion; T cell exhaustion; colorectal cancer; conventional NK cells; immune checkpoints; immune exhaustion genes; innate lymphoid cells.
Copyright © 2021 Sorrentino, D’Antonio, Fieni, Ciummo and Di Carlo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Cytolytic activity correlates with the mutational burden and deregulated expression of immune checkpoints in colorectal cancer.J Exp Clin Cancer Res. 2019 Aug 20;38(1):364. doi: 10.1186/s13046-019-1372-z. J Exp Clin Cancer Res. 2019. PMID: 31429779 Free PMC article.
-
Expression of immune checkpoints and T cell exhaustion markers in early and advanced stages of colorectal cancer.Cancer Immunol Immunother. 2020 Oct;69(10):1989-1999. doi: 10.1007/s00262-020-02593-w. Epub 2020 May 11. Cancer Immunol Immunother. 2020. PMID: 32393998 Free PMC article.
-
Prognostic impacts of tumoral expression and serum levels of PD-L1 and CTLA-4 in colorectal cancer patients.Cancer Immunol Immunother. 2020 Dec;69(12):2533-2546. doi: 10.1007/s00262-020-02645-1. Epub 2020 Jun 23. Cancer Immunol Immunother. 2020. PMID: 32577816 Free PMC article.
-
The Role of the Immune Infiltrate in Distinct Cancer Types and Its Clinical Implications : Lymphocytic Infiltration in Colorectal Cancer.Cancer Treat Res. 2020;180:197-211. doi: 10.1007/978-3-030-38862-1_7. Cancer Treat Res. 2020. PMID: 32215871 Review.
-
T-cell immunoglobulin and ITIM domain, as a potential immune checkpoint target for immunotherapy of colorectal cancer.IUBMB Life. 2021 May;73(5):726-738. doi: 10.1002/iub.2461. Epub 2021 Mar 30. IUBMB Life. 2021. PMID: 33686787 Review.
Cited by
-
Regulation of CD4 T Cell Responses by the Transcription Factor Eomesodermin.Biomolecules. 2022 Oct 24;12(11):1549. doi: 10.3390/biom12111549. Biomolecules. 2022. PMID: 36358898 Free PMC article. Review.
-
An Analysis Regarding the Association Between DAZ Interacting Zinc Finger Protein 1 (DZIP1) and Colorectal Cancer (CRC).Mol Biotechnol. 2025 Feb;67(2):527-547. doi: 10.1007/s12033-024-01065-1. Epub 2024 Feb 9. Mol Biotechnol. 2025. PMID: 38334905
-
Regulator of telomere elongation helicase 1 gene and its association with malignancy.Cancer Rep (Hoboken). 2023 Jan;6(1):e1735. doi: 10.1002/cnr2.1735. Epub 2022 Oct 17. Cancer Rep (Hoboken). 2023. PMID: 36253342 Free PMC article. Review.
-
Canopy FGF signaling regulator 3 affects prognosis, immune infiltration, and PI3K/AKT pathway in colon adenocarcinoma.World J Gastrointest Oncol. 2024 Jul 15;16(7):3284-3298. doi: 10.4251/wjgo.v16.i7.3284. World J Gastrointest Oncol. 2024. PMID: 39072149 Free PMC article.
-
Crosstalk between colorectal CSCs and immune cells in tumorigenesis, and strategies for targeting colorectal CSCs.Exp Hematol Oncol. 2024 Jan 22;13(1):6. doi: 10.1186/s40164-024-00474-x. Exp Hematol Oncol. 2024. PMID: 38254219 Free PMC article. Review.
References
-
- World Health Organization . Cancer Today (2020). Available at: https://gco.iarc.fr/today/home (Accessed September 02, 2021).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

