Fragment-Based Ligand Discovery Applied to the Mycolic Acid Methyltransferase Hma (MmaA4) from Mycobacterium tuberculosis: A Crystallographic and Molecular Modelling Study
- PMID: 34959681
- PMCID: PMC8708032
- DOI: 10.3390/ph14121282
Fragment-Based Ligand Discovery Applied to the Mycolic Acid Methyltransferase Hma (MmaA4) from Mycobacterium tuberculosis: A Crystallographic and Molecular Modelling Study
Abstract
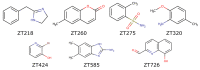
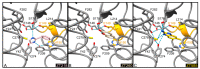
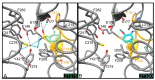
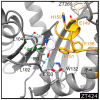
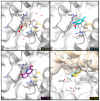
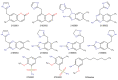
The mycolic acid biosynthetic pathway represents a promising source of pharmacological targets in the fight against tuberculosis. In Mycobacterium tuberculosis, mycolic acids are subject to specific chemical modifications introduced by a set of eight S-adenosylmethionine dependent methyltransferases. Among these, Hma (MmaA4) is responsible for the introduction of oxygenated modifications. Crystallographic screening of a library of fragments allowed the identification of seven ligands of Hma. Two mutually exclusive binding modes were identified, depending on the conformation of residues 147-154. These residues are disordered in apo-Hma but fold upon binding of the S-adenosylmethionine (SAM) cofactor as well as of analogues, resulting in the formation of the short η1-helix. One of the observed conformations would be incompatible with the presence of the cofactor, suggesting that allosteric inhibitors could be designed against Hma. Chimeric compounds were designed by fusing some of the bound fragments, and the relative binding affinities of initial fragments and evolved compounds were investigated using molecular dynamics simulation and generalised Born and Poisson-Boltzmann calculations coupled to the surface area continuum solvation method. Molecular dynamics simulations were also performed on apo-Hma to assess the structural plasticity of the unliganded protein. Our results indicate a significant improvement in the binding properties of the designed compounds, suggesting that they could be further optimised to inhibit Hma activity.
Keywords: Mycobacterium tuberculosis; binding energies; fragment-based ligand discovery; molecular modelling; mycolic acid methyltransferases.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
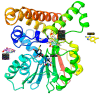

Similar articles
-
Further insight into S-adenosylmethionine-dependent methyltransferases: structural characterization of Hma, an enzyme essential for the biosynthesis of oxygenated mycolic acids in Mycobacterium tuberculosis.J Biol Chem. 2006 Feb 17;281(7):4434-45. doi: 10.1074/jbc.M510250200. Epub 2005 Dec 15. J Biol Chem. 2006. PMID: 16356931
-
S-adenosyl-N-decyl-aminoethyl, a potent bisubstrate inhibitor of mycobacterium tuberculosis mycolic acid methyltransferases.J Biol Chem. 2009 Jul 17;284(29):19321-30. doi: 10.1074/jbc.M809599200. Epub 2009 May 13. J Biol Chem. 2009. PMID: 19439410 Free PMC article.
-
Mycolic acid modification by the mmaA4 gene of M. tuberculosis modulates IL-12 production.PLoS Pathog. 2008 Jun 6;4(6):e1000081. doi: 10.1371/journal.ppat.1000081. PLoS Pathog. 2008. PMID: 18535659 Free PMC article.
-
New approaches to target the mycolic acid biosynthesis pathway for the development of tuberculosis therapeutics.Curr Pharm Des. 2014;20(27):4357-78. doi: 10.2174/1381612819666131118203641. Curr Pharm Des. 2014. PMID: 24245756 Free PMC article. Review.
-
Drugs that inhibit mycolic acid biosynthesis in Mycobacterium tuberculosis.Curr Pharm Biotechnol. 2002 Sep;3(3):197-225. doi: 10.2174/1389201023378328. Curr Pharm Biotechnol. 2002. PMID: 12164478 Review.
Cited by
-
The pathogenic mechanism of Mycobacterium tuberculosis: implication for new drug development.Mol Biomed. 2022 Dec 22;3(1):48. doi: 10.1186/s43556-022-00106-y. Mol Biomed. 2022. PMID: 36547804 Free PMC article. Review.
References
-
- World Health Organization . Global Tuberculosis Report 2021. World Health Organization; Geneva, Switzerland: 2021.
Grants and funding
LinkOut - more resources
Full Text Sources

