NMR Hydrophilic Metabolomic Analysis of Bacterial Resistance Pathways Using Multivalent Antimicrobials with Challenged and Unchallenged Wild Type and Mutated Gram-Positive Bacteria
- PMID: 34948402
- PMCID: PMC8715671
- DOI: 10.3390/ijms222413606
NMR Hydrophilic Metabolomic Analysis of Bacterial Resistance Pathways Using Multivalent Antimicrobials with Challenged and Unchallenged Wild Type and Mutated Gram-Positive Bacteria
Abstract
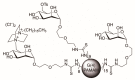
Multivalent membrane disruptors are a relatively new antimicrobial scaffold that are difficult for bacteria to develop resistance to and can act on both Gram-positive and Gram-negative bacteria. Proton Nuclear Magnetic Resonance (1H NMR) metabolomics is an important method for studying resistance development in bacteria, since this is both a quantitative and qualitative method to study and identify phenotypes by changes in metabolic pathways. In this project, the metabolic differences between wild type Bacillus cereus (B. cereus) samples and B. cereus that was mutated through 33 growth cycles in a nonlethal dose of a multivalent antimicrobial agent were identified. For additional comparison, samples for analysis of the wild type and mutated strains of B. cereus were prepared in both challenged and unchallenged conditions. A C16-DABCO (1,4-diazabicyclo-2,2,2-octane) and mannose functionalized poly(amidoamine) dendrimer (DABCOMD) were used as the multivalent quaternary ammonium antimicrobial for this hydrophilic metabolic analysis. Overall, the study reported here indicates that B. cereus likely change their peptidoglycan layer to protect themselves from the highly positively charged DABCOMD. This membrane fortification most likely leads to the slow growth curve of the mutated, and especially the challenged mutant samples. The association of these sample types with metabolites associated with energy expenditure is attributed to the increased energy required for the membrane fortifications to occur as well as to the decreased diffusion of nutrients across the mutated membrane.
Keywords: Bacillus cereus; DABCO; Gram-positive bacteria; antibiotic resistance; dendrimers; membrane disruption; metabolomics; nuclear magnetic resonance; quaternary ammonium compounds.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




Similar articles
-
NMR metabolomic analysis of bacterial resistance pathways using multivalent quaternary ammonium functionalized macromolecules.Metabolomics. 2020 Jul 23;16(8):82. doi: 10.1007/s11306-020-01702-1. Metabolomics. 2020. PMID: 32705355 Free PMC article.
-
Synthesis and Biological Activity of Highly Cationic Dendrimer Antibiotics.Mol Pharm. 2016 Nov 7;13(11):3827-3834. doi: 10.1021/acs.molpharmaceut.6b00628. Epub 2016 Oct 4. Mol Pharm. 2016. PMID: 27661609 Free PMC article.
-
Evaluation of two transformation protocols and screening of positive plasmid introduction into Bacillus cereus EB2, a gram-positive bacterium using qualitative analyses.Braz J Microbiol. 2020 Sep;51(3):919-929. doi: 10.1007/s42770-020-00241-0. Epub 2020 Feb 20. Braz J Microbiol. 2020. PMID: 32078730 Free PMC article.
-
Evolving resistance among Gram-positive pathogens.Clin Infect Dis. 2015 Sep 15;61 Suppl 2(Suppl 2):S48-57. doi: 10.1093/cid/civ523. Clin Infect Dis. 2015. PMID: 26316558 Free PMC article. Review.
-
Mechanisms of drug resistance: daptomycin resistance.Ann N Y Acad Sci. 2015 Sep;1354:32-53. doi: 10.1111/nyas.12948. Epub 2015 Oct 23. Ann N Y Acad Sci. 2015. PMID: 26495887 Free PMC article. Review.
Cited by
-
Metabolomic Profiles of Multidrug-Resistant Salmonella Typhimurium from Humans, Bovine, and Porcine Hosts.Animals (Basel). 2022 Jun 10;12(12):1518. doi: 10.3390/ani12121518. Animals (Basel). 2022. PMID: 35739855 Free PMC article.
-
The Integration of Proteomics and Metabolomics Data Paving the Way for a Better Understanding of the Mechanisms Underlying Microbial Acquired Drug Resistance.Front Med (Lausanne). 2022 May 6;9:849838. doi: 10.3389/fmed.2022.849838. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35602483 Free PMC article.
References
-
- Todar K. Textbook of Bacteriology. University of Wisconsin; Madison, WI, USA: 2020. [(accessed on 10 November 2021)]. Available online: http://textbookofbacteriology.net.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

