Genome Features of a New Double-Stranded RNA Helper Virus (LBCbarr) from Wine Torulaspora delbrueckii Killer Strains
- PMID: 34948288
- PMCID: PMC8709356
- DOI: 10.3390/ijms222413492
Genome Features of a New Double-Stranded RNA Helper Virus (LBCbarr) from Wine Torulaspora delbrueckii Killer Strains
Abstract
The killer phenotype of Torulaspora delbrueckii (Td) and Saccharomyces cerevisiae (Sc) is encoded in the genome of medium-size dsRNA viruses (V-M). Killer strains also contain a helper large size (4.6 kb) dsRNA virus (V-LA) which is required for maintenance and replication of V-M. Another large-size (4.6 kb) dsRNA virus (V-LBC), without known helper activity to date, may join V-LA and V-M in the same yeast. T. delbrueckii Kbarr1 killer strain contains the killer virus Mbarr1 in addition to two L viruses, TdV-LAbarr1 and TdV-LBCbarr1. In contrast, the T. delbrueckii Kbarr2 killer strain contains two M killer viruses (Mbarr1 and M1) and a LBC virus (TdV-LBCbarr2), which has helper capability to maintain both M viruses. The genomes of TdV-LBCbarr1 and TdV-LBCbarr2 were characterized by high-throughput sequencing (HTS). Both RNA genomes share sequence identity and similar organization with their ScV-LBC counterparts. They contain all conserved motifs required for translation, packaging, and replication of viral RNA. Their Gag-Pol amino-acid sequences also contain the features required for cap-snatching and RNA polymerase activity. However, some of these motifs and features are similar to those of LA viruses, which may explain that at least TdV-LBCbarr2 has a helper ability to maintain M killer viruses. Newly sequenced ScV-LBC genomes contained the same motifs and features previously found in LBC viruses, with the same genome location and secondary structure. Sequence comparison showed that LBC viruses belong to two clusters related to each species of yeast. No evidence for associated co-evolution of specific LBC with specific M virus was found. The presence of the same M1 virus in S. cerevisiae and T. delbrueckii raises the possibility of cross-species transmission of M viruses.
Keywords: LBC helper virus; Torulaspora delbrueckii; dsRNA genome; high-throughput sequencing; killer Kbarr.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

 , ribosomal frameshift. Asterisks (*) indicate identical nucleotide positions. The secondary structures of the putative cis signals for frameshifting, packaging, and replication of TdV-LBCbarr2 are displayed at the right of the sequence panel.
, ribosomal frameshift. Asterisks (*) indicate identical nucleotide positions. The secondary structures of the putative cis signals for frameshifting, packaging, and replication of TdV-LBCbarr2 are displayed at the right of the sequence panel.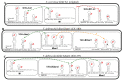
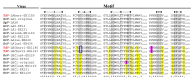
 , ribosomal frameshift. Black arrow indicates the single nucleotide changes between ES of LBCbarr1 and LBCbarr2. Green dot-line arrow indicates equivalent ES for L and M viruses in the same yeast. Orange dot-line arrow indicates possible equivalent ES for LBCbarr1 and Mbarr1 in Kbarr1 EX1180 yeast. Red dot-line arrow indicates unknown equivalent ES in M viruses for the ES of ScV-LBC1-original.
, ribosomal frameshift. Black arrow indicates the single nucleotide changes between ES of LBCbarr1 and LBCbarr2. Green dot-line arrow indicates equivalent ES for L and M viruses in the same yeast. Orange dot-line arrow indicates possible equivalent ES for LBCbarr1 and Mbarr1 in Kbarr1 EX1180 yeast. Red dot-line arrow indicates unknown equivalent ES in M viruses for the ES of ScV-LBC1-original.

Similar articles
-
New Insights into the Genome Organization of Yeast Double-Stranded RNA LBC Viruses.Microorganisms. 2022 Jan 14;10(1):173. doi: 10.3390/microorganisms10010173. Microorganisms. 2022. PMID: 35056622 Free PMC article.
-
Genome Organization of a New Double-Stranded RNA LA Helper Virus From Wine Torulaspora delbrueckii Killer Yeast as Compared With Its Saccharomyces Counterparts.Front Microbiol. 2020 Nov 23;11:593846. doi: 10.3389/fmicb.2020.593846. eCollection 2020. Front Microbiol. 2020. PMID: 33324373 Free PMC article.
-
New Insights into the Genome Organization of Yeast Killer Viruses Based on "Atypical" Killer Strains Characterized by High-Throughput Sequencing.Toxins (Basel). 2017 Sep 19;9(9):292. doi: 10.3390/toxins9090292. Toxins (Basel). 2017. PMID: 28925975 Free PMC article.
-
Double-stranded and single-stranded RNA viruses of Saccharomyces cerevisiae.Annu Rev Microbiol. 1992;46:347-75. doi: 10.1146/annurev.mi.46.100192.002023. Annu Rev Microbiol. 1992. PMID: 1444259 Review.
-
Yeast dsRNA viruses: replication and killer phenotypes.Mol Microbiol. 1991 Oct;5(10):2331-8. doi: 10.1111/j.1365-2958.1991.tb02078.x. Mol Microbiol. 1991. PMID: 1665194 Review.
Cited by
-
New Insights into the Genome Organization of Yeast Double-Stranded RNA LBC Viruses.Microorganisms. 2022 Jan 14;10(1):173. doi: 10.3390/microorganisms10010173. Microorganisms. 2022. PMID: 35056622 Free PMC article.
-
The Life of Saccharomyces and Non-Saccharomyces Yeasts in Drinking Wine.Microorganisms. 2023 Apr 30;11(5):1178. doi: 10.3390/microorganisms11051178. Microorganisms. 2023. PMID: 37317152 Free PMC article. Review.
-
The evolutionary ecology of fungal killer phenotypes.Proc Biol Sci. 2023 Aug 30;290(2005):20231108. doi: 10.1098/rspb.2023.1108. Epub 2023 Aug 16. Proc Biol Sci. 2023. PMID: 37583325 Free PMC article. Review.
-
Adaptive Response of Saccharomyces Hosts to Totiviridae L-A dsRNA Viruses Is Achieved through Intrinsically Balanced Action of Targeted Transcription Factors.J Fungi (Basel). 2022 Apr 9;8(4):381. doi: 10.3390/jof8040381. J Fungi (Basel). 2022. PMID: 35448612 Free PMC article.
-
Biocontrol Using Torulaspora delbrueckii in Sequential Fermentation: New Insights into Low-Sulfite Verdicchio Wines.Foods. 2023 Jul 30;12(15):2899. doi: 10.3390/foods12152899. Foods. 2023. PMID: 37569169 Free PMC article.
References
-
- Rodríguez-Cousiño N., Maqueda M., Ambrona J., Zamora E., Esteban E., Ramírez M. A new wine Saccharomyces cerevisiae double-stranded RNA virus encoded killer toxin (Klus) with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl. Environ. Microbiol. 2011;77:1822–1832. doi: 10.1128/AEM.02501-10. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

