Protein Receptors Evolved from Homologous Cohesion Modules That Self-Associated and Are Encoded by Interactive Networked Genes
- PMID: 34947866
- PMCID: PMC8707797
- DOI: 10.3390/life11121335
Protein Receptors Evolved from Homologous Cohesion Modules That Self-Associated and Are Encoded by Interactive Networked Genes
Abstract
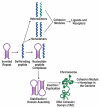
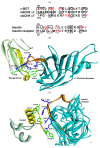
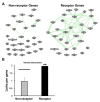
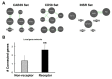
Previously, it was proposed that protein receptors evolved from self-binding peptides that were encoded by self-interacting gene segments (inverted repeats) widely dispersed in the genome. In addition, self-association of the peptides was thought to be mediated by regions of amino acid sequence similarity. To extend these ideas, special features of receptors have been explored, such as their degree of homology to other proteins, and the arrangement of their genes for clues about their evolutionary origins and dynamics in the genome. As predicted, BLASTP searches for homologous proteins detected a greater number of unique hits for queries with receptor sequences than for sequences of randomly-selected, non-receptor proteins. This suggested that the building blocks (cohesion modules) for receptors were duplicated, dispersed, and maintained in the genome, due to structure/function relationships discussed here. Furthermore, the genes coding for a representative panel of receptors participated in a larger number of gene-gene interactions than for randomly-selected genes. This could conceivably reflect a greater evolutionary conservation of the receptor genes, with their more extensive integration into networks along with inherent properties of the genes themselves. In support of the latter possibility, some receptor genes were located in active areas of adaptive gene relocation/amalgamation to form functional blocks of related genes. It is suggested that adaptive relocation might allow for their joint regulation by common promoters and enhancers, and affect local chromatin structural domains to facilitate or repress gene expression. Speculation is included about the nature of the coordinated communication between receptors and the genes that encode them.
Keywords: cohesion modules; gene interaction networks; receptor evolution; self-organization; syntenic blocks of genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Macromolecular crowding: chemistry and physics meet biology (Ascona, Switzerland, 10-14 June 2012).Phys Biol. 2013 Aug;10(4):040301. doi: 10.1088/1478-3975/10/4/040301. Epub 2013 Aug 2. Phys Biol. 2013. PMID: 23912807
-
Assembly of exons from unitary transposable genetic elements: implications for the evolution of protein-protein interactions.J Theor Biol. 1998 Sep 7;194(1):11-27. doi: 10.1006/jtbi.1998.0676. J Theor Biol. 1998. PMID: 9778422
-
Engineering Aspects of Olfaction.In: Persaud KC, Marco S, Gutiérrez-Gálvez A, editors. Neuromorphic Olfaction. Boca Raton (FL): CRC Press/Taylor & Francis; 2013. Chapter 1. In: Persaud KC, Marco S, Gutiérrez-Gálvez A, editors. Neuromorphic Olfaction. Boca Raton (FL): CRC Press/Taylor & Francis; 2013. Chapter 1. PMID: 26042329 Free Books & Documents. Review.
-
The ribosome as a missing link in prebiotic evolution II: Ribosomes encode ribosomal proteins that bind to common regions of their own mRNAs and rRNAs.J Theor Biol. 2016 May 21;397:115-27. doi: 10.1016/j.jtbi.2016.02.030. Epub 2016 Mar 4. J Theor Biol. 2016. PMID: 26953650
-
The ancestry and cumulative evolution of immune reactions.Acta Biochim Pol. 2010;57(4):443-66. Epub 2010 Nov 1. Acta Biochim Pol. 2010. PMID: 21046016 Review.
References
LinkOut - more resources
Full Text Sources
Research Materials

