Mitophagy in Yeast: Decades of Research
- PMID: 34944049
- PMCID: PMC8700663
- DOI: 10.3390/cells10123541
Mitophagy in Yeast: Decades of Research
Abstract
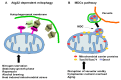
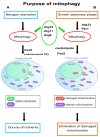
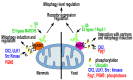
Mitophagy, the selective degradation of mitochondria by autophagy, is one of the most important mechanisms of mitochondrial quality control, and its proper functioning is essential for cellular homeostasis. In this review, we describe the most important milestones achieved during almost 2 decades of research on yeasts, which shed light on the molecular mechanisms, regulation, and role of the Atg32 receptor in this process. We analyze the role of ROS in mitophagy and discuss the physiological roles of mitophagy in unicellular organisms, such as yeast; these roles are very different from those in mammals. Additionally, we discuss some of the different tools available for studying mitophagy.
Keywords: Atg32 protein; mitochondria; mitophagy; quality control; yeast.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Mitophagy in yeast: Molecular mechanisms and physiological role.Biochim Biophys Acta. 2015 Oct;1853(10 Pt B):2756-65. doi: 10.1016/j.bbamcr.2015.01.005. Epub 2015 Jan 17. Biochim Biophys Acta. 2015. PMID: 25603537 Review.
-
Atg32 is a tag for mitochondria degradation in yeast.Autophagy. 2009 Nov;5(8):1201-2. doi: 10.4161/auto.5.8.9747. Epub 2009 Nov 6. Autophagy. 2009. PMID: 19736522
-
Mitophagy in Yeast: Molecular Mechanism and Regulation.Cells. 2021 Dec 17;10(12):3569. doi: 10.3390/cells10123569. Cells. 2021. PMID: 34944077 Free PMC article. Review.
-
Mechanisms and Physiological Roles of Mitophagy in Yeast.Mol Cells. 2018 Jan 31;41(1):35-44. doi: 10.14348/molcells.2018.2214. Epub 2018 Jan 23. Mol Cells. 2018. PMID: 29370687 Free PMC article. Review.
-
Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast.J Biol Chem. 2012 Jan 27;287(5):3265-72. doi: 10.1074/jbc.M111.280156. Epub 2011 Dec 7. J Biol Chem. 2012. PMID: 22157017 Free PMC article.
Cited by
-
Current opinions on mitophagy in fungi.Autophagy. 2023 Mar;19(3):747-757. doi: 10.1080/15548627.2022.2098452. Epub 2022 Jul 11. Autophagy. 2023. PMID: 35793406 Free PMC article. Review.
-
Ectopic BH3-Only Protein Bim Associates with Hsp70 to Regulate Yeast Mitophagy.Dokl Biochem Biophys. 2023 Oct;512(1):292-299. doi: 10.1134/S1607672923700485. Epub 2023 Dec 13. Dokl Biochem Biophys. 2023. PMID: 38093134 Free PMC article.
-
β-Cyclocitral from Lavandula angustifolia Mill. Exerts Anti-Aging Effects on Yeasts and Mammalian Cells via Telomere Protection, Antioxidative Stress, and Autophagy Activation.Antioxidants (Basel). 2024 Jun 12;13(6):715. doi: 10.3390/antiox13060715. Antioxidants (Basel). 2024. PMID: 38929154 Free PMC article.
-
Mitochondrial Glutathione in Cellular Redox Homeostasis and Disease Manifestation.Int J Mol Sci. 2024 Jan 21;25(2):1314. doi: 10.3390/ijms25021314. Int J Mol Sci. 2024. PMID: 38279310 Free PMC article. Review.
References
-
- Chance B. The respiratory chain and oxidative phosphorylation. Adv. Enzymol. Relat. Subj. Biochem. 1956;17:65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

