Metagenomic Analysis of the Pediatric-Onset Multiple Sclerosis Gut Microbiome
- PMID: 34937787
- PMCID: PMC8967388
- DOI: 10.1212/WNL.0000000000013245
Metagenomic Analysis of the Pediatric-Onset Multiple Sclerosis Gut Microbiome
Abstract
Background and objectives: Little is known of the functional potential of the gut microbiome in pediatric-onset multiple sclerosis (MS). We performed metagenomic analyses using stool samples from individuals with pediatric-onset MS and unaffected controls.
Methods: Persons ≤21 years old enrolled in the Canadian Pediatric Demyelinating Disease Network providing a stool sample were eligible. Twenty patients with MS (McDonald criteria) with symptom onset <18 years were matched to 20 controls by sex, age (±3 years), stool consistency, and race. Microbial taxonomy and functional potentials were estimated from stool sample-derived metagenomic reads and compared by disease status (MS vs controls) and disease-modifying drug (DMD) exposure using alpha diversity, relative abundance, and prevalence using Wilcoxon rank sum, ALDEx2, and Fisher exact tests, respectively.
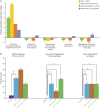
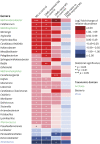
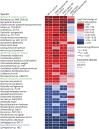
Results: Individuals with MS were aged 13.6 years (mean) at symptom onset and 8 were DMD-naive. Mean ages at stool sample were 16.1 and 15.4 years for MS and control participants, respectively; 80% were girls. Alpha diversity of enzymes and proteins did not differ by disease or DMD status (p > 0.20), but metabolic pathways, gene annotations, and microbial taxonomy did. Individuals with MS (vs controls) exhibited higher methanogenesis prevalence (odds ratio 10, p = 0.044) and Methanobrevibacter abundance (log2 fold change [LFC] 1.7, p = 0.0014), but lower homolactic fermentation abundance (LFC -0.48, p = 0.039). Differences by DMD status included lower phosphate butyryl transferase for DMD-naive vs exposed patients with MS (LFC -1.0, p = 0.033).
Discussion: The gut microbiome's functional potential and taxonomy differed between individuals with pediatric-onset MS vs controls, including higher prevalence of a methane-producing pathway from Archaea and depletion of the lactate fermentation pathway. DMD exposure was associated with butyrate-producing enzyme enrichment. Together these findings indicate that the gut microbiome of individuals with MS may have a disturbed functional potential.
© 2021 American Academy of Neurology.
Figures


Similar articles
-
The metabolic potential of the paediatric-onset multiple sclerosis gut microbiome.Mult Scler Relat Disord. 2022 Jul;63:103829. doi: 10.1016/j.msard.2022.103829. Epub 2022 Apr 23. Mult Scler Relat Disord. 2022. PMID: 35500534
-
The gut microbiota in pediatric multiple sclerosis and demyelinating syndromes.Ann Clin Transl Neurol. 2021 Dec;8(12):2252-2269. doi: 10.1002/acn3.51476. Epub 2021 Dec 9. Ann Clin Transl Neurol. 2021. PMID: 34889081 Free PMC article.
-
A cross-sectional study of MRI features and the gut microbiome in pediatric-onset multiple sclerosis.Ann Clin Transl Neurol. 2024 Feb;11(2):486-496. doi: 10.1002/acn3.51970. Epub 2023 Dec 21. Ann Clin Transl Neurol. 2024. PMID: 38130033 Free PMC article.
-
Gut microbiome research in multiple sclerosis.Neurosci Res. 2021 Jul;168:28-31. doi: 10.1016/j.neures.2021.05.001. Epub 2021 May 11. Neurosci Res. 2021. PMID: 33989681 Review.
-
Gut microbiome in multiple sclerosis: The players involved and the roles they play.Gut Microbes. 2017 Nov 2;8(6):607-615. doi: 10.1080/19490976.2017.1349041. Epub 2017 Aug 3. Gut Microbes. 2017. PMID: 28696139 Free PMC article. Review.
Cited by
-
Association of nutritional intake with clinical and imaging activity in pediatric multiple sclerosis.Mult Scler. 2024 Jul;30(8):1056-1065. doi: 10.1177/13524585241261556. Mult Scler. 2024. PMID: 39078111 Free PMC article.
-
Correlation between the gut microbiome and neurodegenerative diseases: a review of metagenomics evidence.Neural Regen Res. 2024 Apr;19(4):833-845. doi: 10.4103/1673-5374.382223. Neural Regen Res. 2024. PMID: 37843219 Free PMC article. Review.
-
Environmental Influences on Risk and Disease Course in Pediatric Multiple Sclerosis.Semin Pediatr Neurol. 2023 Jul;46:101049. doi: 10.1016/j.spen.2023.101049. Epub 2023 Apr 25. Semin Pediatr Neurol. 2023. PMID: 37451747 Free PMC article.
-
Mediterranean diet and associations with the gut microbiota and pediatric-onset multiple sclerosis using trivariate analysis.Commun Med (Lond). 2024 Jul 19;4(1):148. doi: 10.1038/s43856-024-00565-0. Commun Med (Lond). 2024. PMID: 39030379 Free PMC article.
-
The interplay between the gut-brain axis and the microbiome: A perspective on psychiatric and neurodegenerative disorders.Front Neurosci. 2022 Oct 28;16:1030694. doi: 10.3389/fnins.2022.1030694. eCollection 2022. Front Neurosci. 2022. PMID: 36389228 Free PMC article. Review.
References
-
- Thompson AJ, Baranzini SE, Geurts J, Hemmer B, Ciccarelli O. Multiple sclerosis. Lancet. 2018;391(10130):1622-1636. - PubMed
-
- Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19(1):55-71. - PubMed
-
- Tremlett H, Bauer KC, Appel-Cresswell S, Finlay BB, Waubant E. The gut microbiome in human neurological disease: a review. Ann Neurol. 2017;81(3):369-382. - PubMed
