Amyloid-like aggregating proteins cause lysosomal defects in neurons via gain-of-function toxicity
- PMID: 34933920
- PMCID: PMC8711852
- DOI: 10.26508/lsa.202101185
Amyloid-like aggregating proteins cause lysosomal defects in neurons via gain-of-function toxicity
Abstract
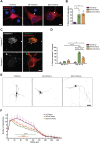
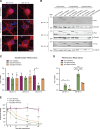
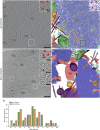
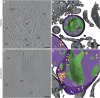
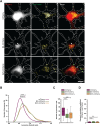
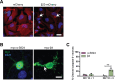
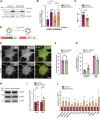
The autophagy-lysosomal pathway is impaired in many neurodegenerative diseases characterized by protein aggregation, but the link between aggregation and lysosomal dysfunction remains poorly understood. Here, we combine cryo-electron tomography, proteomics, and cell biology studies to investigate the effects of protein aggregates in primary neurons. We use artificial amyloid-like β-sheet proteins (β proteins) to focus on the gain-of-function aspect of aggregation. These proteins form fibrillar aggregates and cause neurotoxicity. We show that late stages of autophagy are impaired by the aggregates, resulting in lysosomal alterations reminiscent of lysosomal storage disorders. Mechanistically, β proteins interact with and sequester AP-3 μ1, a subunit of the AP-3 adaptor complex involved in protein trafficking to lysosomal organelles. This leads to destabilization of the AP-3 complex, missorting of AP-3 cargo, and lysosomal defects. Restoring AP-3μ1 expression ameliorates neurotoxicity caused by β proteins. Altogether, our results highlight the link between protein aggregation, lysosomal impairments, and neurotoxicity.
© 2021 Riera-Tur et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Autoimmune Responses to Soluble Aggregates of Amyloidogenic Proteins Involved in Neurodegenerative Diseases: Overlapping Aggregation Prone and Autoimmunogenic regions.Sci Rep. 2016 Feb 29;6:22258. doi: 10.1038/srep22258. Sci Rep. 2016. PMID: 26924748 Free PMC article.
-
Gallic acid oxidation products alter the formation pathway of insulin amyloid fibrils.Sci Rep. 2020 Sep 2;10(1):14466. doi: 10.1038/s41598-020-70982-3. Sci Rep. 2020. PMID: 32879381 Free PMC article.
-
Loss of Cathepsin B and L Leads to Lysosomal Dysfunction, NPC-Like Cholesterol Sequestration and Accumulation of the Key Alzheimer's Proteins.PLoS One. 2016 Nov 30;11(11):e0167428. doi: 10.1371/journal.pone.0167428. eCollection 2016. PLoS One. 2016. PMID: 27902765 Free PMC article.
-
Autophagy Modulation as a Treatment of Amyloid Diseases.Molecules. 2019 Sep 16;24(18):3372. doi: 10.3390/molecules24183372. Molecules. 2019. PMID: 31527516 Free PMC article. Review.
-
Single Molecule Characterization of Amyloid Oligomers.Molecules. 2021 Feb 11;26(4):948. doi: 10.3390/molecules26040948. Molecules. 2021. PMID: 33670093 Free PMC article. Review.
Cited by
-
Gel-like inclusions of C-terminal fragments of TDP-43 sequester stalled proteasomes in neurons.EMBO Rep. 2022 Jun 7;23(6):e53890. doi: 10.15252/embr.202153890. Epub 2022 Apr 19. EMBO Rep. 2022. PMID: 35438230 Free PMC article.
-
Handling Difficult Cryo-ET Samples: A Study with Primary Neurons from Drosophila melanogaster.Microsc Microanal. 2023 Dec 21;29(6):2127-2148. doi: 10.1093/micmic/ozad125. Microsc Microanal. 2023. PMID: 37966978 Free PMC article.
-
A cryo-ET survey of microtubules and intracellular compartments in mammalian axons.J Cell Biol. 2022 Feb 7;221(2):e202103154. doi: 10.1083/jcb.202103154. Epub 2021 Dec 8. J Cell Biol. 2022. PMID: 34878519 Free PMC article.
-
Mechanisms and pathology of protein misfolding and aggregation.Nat Rev Mol Cell Biol. 2023 Dec;24(12):912-933. doi: 10.1038/s41580-023-00647-2. Epub 2023 Sep 8. Nat Rev Mol Cell Biol. 2023. PMID: 37684425 Review.
-
Therapeutic effects of a novel synthetic α-secretase.Front Aging Neurosci. 2024 Jun 7;16:1383905. doi: 10.3389/fnagi.2024.1383905. eCollection 2024. Front Aging Neurosci. 2024. PMID: 38912519 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
