Comparative Evaluation of the Ileum Microbiota Composition in Piglets at Different Growth Stages
- PMID: 34925272
- PMCID: PMC8672721
- DOI: 10.3389/fmicb.2021.765691
Comparative Evaluation of the Ileum Microbiota Composition in Piglets at Different Growth Stages
Abstract
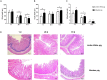
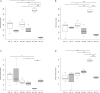
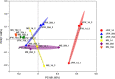
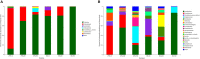
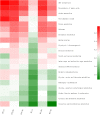
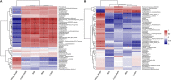
Intestinal microbiota can affect the intake, storage, and absorption of nutrients in the body, thereby greatly impacting the growth and development of animals. In addition to diet, the breed and growth stages of pigs could also affect changes in the intestinal microbiota. However, research on the developmental changes in the ileum microbiota of piglets remains unclear. In this study, the ileum microbiota of Jinfen White and Mashen piglets at different developmental stages were investigated using 16S rRNA sequencing. Physiologically, the villus height of the ileum decreased, and the crypt depth increased during the development of the two pig breeds. Additionally, the serum antioxidant factors in the Jinfen White piglets were significantly higher than in the Mashen piglets at the end of the nursing stage. A total of 690 operational taxonomic units (OTUs) belonging to 21 phyla and 286 genera were identified, of which Firmicutes and Proteobacteria were the dominant phyla during the development of both the Jinfen White and Mashen piglets, accounting for ∼90% of all OTUs. Further research revealed differences in dominant bacteria between the two breeds. With increasing age, the ileum microbial diversity increased, and in both the pig breeds, the proportion of Firmicutes increased, whereas the proportion of Proteobacteria decreased. Additionally, different samples were characterized by specific genera, and different Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were predicted at certain developmental stages. Finally, the correlation between the ileum microbiota and physiological features was analyzed, and it was suggested that the host and environmental factors play important roles in the formation of the microbial community structure in piglets. In summary, we delineated the structure, function, and differences in ileum microbiota between Jinfen White and Mashen piglets during different growth stages. This study helps to understand the development of the intestinal microbiota in local and hybrid pig breeds.
Keywords: 16S rRNA; Jinfen White and Mashen piglets; growth stage; ileum microbiota; intestinal microorganism.
Copyright © 2021 Lu, Liu, Ma, Wang, Cai, Yang, Zhao, Liang, Cao, Li, Kim, Guo and Gao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
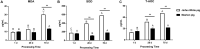

Similar articles
-
Composition of the Fecal Microbiota of Piglets at Various Growth Stages.Front Vet Sci. 2021 Jul 15;8:661671. doi: 10.3389/fvets.2021.661671. eCollection 2021. Front Vet Sci. 2021. PMID: 34336969 Free PMC article.
-
Effects of Maternal Factors and Postpartum Environment on Early Colonization of Intestinal Microbiota in Piglets.Front Vet Sci. 2022 Apr 7;9:815944. doi: 10.3389/fvets.2022.815944. eCollection 2022. Front Vet Sci. 2022. PMID: 35464386 Free PMC article.
-
A comparison of dynamic distributions of intestinal microbiota between Large White and Chinese Shanxi Black pigs.Arch Microbiol. 2019 Apr;201(3):357-367. doi: 10.1007/s00203-019-01620-4. Epub 2019 Jan 23. Arch Microbiol. 2019. PMID: 30673796
-
Gut microbiota of newborn piglets with intrauterine growth restriction have lower diversity and different taxonomic abundances.J Appl Microbiol. 2019 Aug;127(2):354-369. doi: 10.1111/jam.14304. Epub 2019 Jun 7. J Appl Microbiol. 2019. PMID: 31077497 Free PMC article.
-
Fecal microbial composition and functional diversity of Wuzhishan pigs at different growth stages.AMB Express. 2021 Jun 12;11(1):88. doi: 10.1186/s13568-021-01249-x. AMB Express. 2021. PMID: 34117938 Free PMC article.
Cited by
-
Transcriptome Analysis Revealed Potential Genes of Skeletal Muscle Thermogenesis in Mashen Pigs and Large White Pigs under Cold Stress.Int J Mol Sci. 2023 Oct 24;24(21):15534. doi: 10.3390/ijms242115534. Int J Mol Sci. 2023. PMID: 37958518 Free PMC article.
-
Dynamic Changes in the Gut Microbial Community and Function during Broiler Growth.Microbiol Spectr. 2022 Aug 31;10(4):e0100522. doi: 10.1128/spectrum.01005-22. Epub 2022 Aug 11. Microbiol Spectr. 2022. PMID: 35950773 Free PMC article.
-
Effects of Low-Ambient-Temperature Stimulation on Modifying the Intestinal Structure and Function of Different Pig Breeds.Animals (Basel). 2022 Oct 12;12(20):2740. doi: 10.3390/ani12202740. Animals (Basel). 2022. PMID: 36290125 Free PMC article.
-
Maternal tributyrin supplementation in late pregnancy and lactation improves offspring immunity, gut microbiota, and diarrhea rate in a sow model.Front Microbiol. 2023 Apr 12;14:1142174. doi: 10.3389/fmicb.2023.1142174. eCollection 2023. Front Microbiol. 2023. PMID: 37168115 Free PMC article.
-
The effects of differential feeding on ileum development, digestive ability and health status of newborn calves.Front Vet Sci. 2023 Sep 8;10:1255122. doi: 10.3389/fvets.2023.1255122. eCollection 2023. Front Vet Sci. 2023. PMID: 37745216 Free PMC article.
References
-
- Chen H., Mao X. B., Che L. Q., Yu B., He J., Yu J., et al. (2014). Impact of fiber types on gut microbiota, gut environment and gut function in fattening pigs. Anim. Feed Sci. Technol. 195 101–111. 10.1016/j.anifeedsci.2014.06.002 - DOI
LinkOut - more resources
Full Text Sources

