The LORF5 Gene Is Non-essential for Replication but Important for Duck Plague Virus Cell-to-Cell Spread Efficiently in Host Cells
- PMID: 34925260
- PMCID: PMC8674210
- DOI: 10.3389/fmicb.2021.744408
The LORF5 Gene Is Non-essential for Replication but Important for Duck Plague Virus Cell-to-Cell Spread Efficiently in Host Cells
Abstract
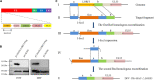
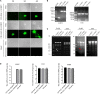
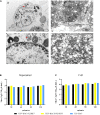
Duck plague virus (DPV) can cause high morbidity and mortality in many waterfowl species within the order Anseriformes. The DPV genome contains 78 open reading frames (ORFs), among which the LORF2, LORF3, LORF4, LORF5, and SORF3 genes are unique genes of avian herpesvirus. In this study, to investigate the role of this unique LORF5 gene in DPV proliferation, we generated a recombinant virus that lacks the LORF5 gene by a two-step red recombination system, which cloned the DPV Chinese virulent strain (DPV CHv) genome into a bacterial artificial chromosome (DPV CHv-BAC); the proliferation law of LORF5-deleted mutant virus on DEF cells and the effect of LORF5 gene on the life cycle stages of DPV compared with the parent strain were tested. Our data revealed that the LORF5 gene contributes to the cell-to-cell transmission of DPV but is not relevant to virus invasion, replication, assembly, and release formation. Taken together, this study sheds light on the role of the avian herpesvirus-specific gene LORF5 in the DPV proliferation life cycle. These findings lay the foundation for in-depth functional studies of the LORF5 gene in DPV or other avian herpesviruses.
Keywords: LORF5 gene; cell-to-cell spread; duck plague virus; non-essential; virus replication.
Copyright © 2021 Shen, Li, Cheng, Wang, Wu, Yang, Jia, Tian, Ou, Mao, Sun, Zhang, Zhu, Chen, Liu, Zhao, Huang, Gao, Liu, Yu, Zhang and Pan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Characterization of a Unique Novel LORF3 Protein of Duck Plague Virus and Its Potential Pathogenesis.J Virol. 2023 Jan 31;97(1):e0157722. doi: 10.1128/jvi.01577-22. Epub 2023 Jan 4. J Virol. 2023. PMID: 36598202 Free PMC article.
-
Duck plague virus gE serves essential functions during the virion final envelopment through influence capsids budding into the cytoplasmic vesicles.Sci Rep. 2020 Mar 27;10(1):5658. doi: 10.1038/s41598-020-62604-9. Sci Rep. 2020. PMID: 32221415 Free PMC article.
-
Duck plague virus Glycoprotein J is functional but slightly impaired in viral replication and cell-to-cell spread.Sci Rep. 2018 Mar 6;8(1):4069. doi: 10.1038/s41598-018-22447-x. Sci Rep. 2018. PMID: 29511274 Free PMC article.
-
Duck virus enteritis (duck plague) - a comprehensive update.Vet Q. 2017 Dec;37(1):57-80. doi: 10.1080/01652176.2017.1298885. Vet Q. 2017. PMID: 28320263 Review.
-
Cloning of herpesviral genomes as bacterial artificial chromosomes.Rev Med Virol. 2003 Mar-Apr;13(2):111-21. doi: 10.1002/rmv.380. Rev Med Virol. 2003. PMID: 12627394 Review.
Cited by
-
Duck plague virus tegument protein vp22 plays a key role in the secondary envelopment and cell-to-cell spread.Vet Res. 2023 Jul 17;54(1):60. doi: 10.1186/s13567-023-01191-9. Vet Res. 2023. PMID: 37461115 Free PMC article.
-
Duck enteritis virus LORF4 gene is a late gene and nonessential for virus replication in vitro.Poult Sci. 2024 Dec;103(12):104275. doi: 10.1016/j.psj.2024.104275. Epub 2024 Aug 31. Poult Sci. 2024. PMID: 39288717 Free PMC article.
-
Characterization of a Unique Novel LORF3 Protein of Duck Plague Virus and Its Potential Pathogenesis.J Virol. 2023 Jan 31;97(1):e0157722. doi: 10.1128/jvi.01577-22. Epub 2023 Jan 4. J Virol. 2023. PMID: 36598202 Free PMC article.
-
Pathogenicity and transmissibility studies on live attenuated duck enteritis virus vaccine in non-target species.Front Microbiol. 2022 Nov 10;13:979368. doi: 10.3389/fmicb.2022.979368. eCollection 2022. Front Microbiol. 2022. PMID: 36439841 Free PMC article.
References
-
- Damiani A. M., Matsumura T., Yokoyama N., Mikami T., Takahashi E. (2000). A deletion in the gI and gE genes of equine herpesvirus type 4 reduces viral virulence in the natural host and affects virus transmission during cell-to-cell spread. Virus Res. 67 189–202. 10.1016/S0168-1702(00)00146-5 - DOI - PubMed
LinkOut - more resources
Full Text Sources

