Seneca Valley virus 3Cpro degrades heterogeneous nuclear ribonucleoprotein A1 to facilitate viral replication
- PMID: 34923914
- PMCID: PMC8923066
- DOI: 10.1080/21505594.2021.2014681
Seneca Valley virus 3Cpro degrades heterogeneous nuclear ribonucleoprotein A1 to facilitate viral replication
Abstract
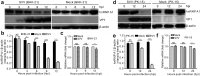
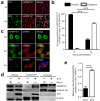
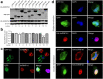
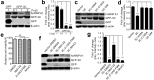
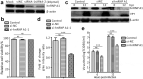
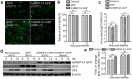
Seneca Valley virus (SVV) is a recently-identified important pathogen that is closely related to idiopathic vesicular disease in swine. Infection of SVV has been shown to induce a variety of cellular factors and their activations are essential for viral replication, but whether heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) involved in SVV replication is unknown. The cytoplasmic redistribution of hnRNP A1 is considered to play an important role in the virus life cycle. Here, we demonstrated that SVV infection can promote redistribution of the nucleocytoplasmic shuttling RNA-binding protein hnRNP A1 to the cytoplasm from the nucleus, whereas hnRNP A1 remained mainly in the nucleus of mock-infected cells. siRNA-mediated knockdown of the gene encoding hnRNP A1 attenuated viral replication as evidenced by decreased viral protein expression and virus production, whereas its overexpression enhanced replication. Moreover, infection with SVV induced the degradation of hnRNP A1, and viral 3 C protease (3 Cpro) was found to be responsible for its degradation and translocation. Further studies demonstrated that 3 Cpro induced hnRNP A1 degradation through its protease activity, via the proteasome pathway. This degradation could be attenuated by a proteasome inhibitor (MG132) and inactivation of the conserved catalytic box in 3 Cpro. Taken together, these results presented here reveal that SVV 3 C protease targets cellular hnRNP A1 for its degradation and translocation, which is utilized by SVV to aid viral replication, thereby highlighting the control potential of strategies for infection of SVV.
Keywords: 3C protease; Seneca valley virus (SVV); degradation; hnRNP A1; replication.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
Similar articles
-
Seneca Valley Virus 3Cpro Mediates Cleavage and Redistribution of Nucleolin To Facilitate Viral Replication.Microbiol Spectr. 2022 Apr 27;10(2):e0030422. doi: 10.1128/spectrum.00304-22. Epub 2022 Mar 31. Microbiol Spectr. 2022. PMID: 35357201 Free PMC article.
-
Seneca Valley Virus 3C pro Cleaves Heterogeneous Nuclear Ribonucleoprotein K to Facilitate Viral Replication.Front Microbiol. 2022 Jul 6;13:945443. doi: 10.3389/fmicb.2022.945443. eCollection 2022. Front Microbiol. 2022. PMID: 35875542 Free PMC article.
-
Seneca valley virus 3C protease blocks EphA2-Mediated mTOR activation to facilitate viral replication.Microb Pathog. 2024 Jun;191:106673. doi: 10.1016/j.micpath.2024.106673. Epub 2024 May 3. Microb Pathog. 2024. PMID: 38705218
-
Seneca Valley virus 3Cpro antagonizes host innate immune responses and programmed cell death.Front Microbiol. 2023 Oct 6;14:1235620. doi: 10.3389/fmicb.2023.1235620. eCollection 2023. Front Microbiol. 2023. PMID: 37869659 Free PMC article. Review.
-
The multifarious roles of heterogeneous ribonucleoprotein A1 in viral infections.Rev Med Virol. 2020 Mar;30(2):e2097. doi: 10.1002/rmv.2097. Epub 2020 Jan 27. Rev Med Virol. 2020. PMID: 31989716 Free PMC article. Review.
Cited by
-
Seneca Valley Virus Induces DHX30 Cleavage to Antagonize Its Antiviral Effects.J Virol. 2022 Sep 14;96(17):e0112122. doi: 10.1128/jvi.01121-22. Epub 2022 Aug 24. J Virol. 2022. PMID: 36000840 Free PMC article.
-
hnRNPA1 impedes snakehead vesiculovirus replication via competitively disrupting viral phosphoprotein-nucleoprotein interaction and degrading viral phosphoprotein.Virulence. 2023 Dec;14(1):2196847. doi: 10.1080/21505594.2023.2196847. Virulence. 2023. PMID: 37005771 Free PMC article.
-
Allosteric regulation of Senecavirus A 3Cpro proteolytic activity by an endogenous phospholipid.PLoS Pathog. 2023 May 30;19(5):e1011411. doi: 10.1371/journal.ppat.1011411. eCollection 2023 May. PLoS Pathog. 2023. PMID: 37253057 Free PMC article.
-
Seneca Valley Virus 3Cpro Mediates Cleavage and Redistribution of Nucleolin To Facilitate Viral Replication.Microbiol Spectr. 2022 Apr 27;10(2):e0030422. doi: 10.1128/spectrum.00304-22. Epub 2022 Mar 31. Microbiol Spectr. 2022. PMID: 35357201 Free PMC article.
-
2B and 3C Proteins of Senecavirus A Antagonize the Antiviral Activity of DDX21 via the Caspase-Dependent Degradation of DDX21.Front Immunol. 2022 Jul 14;13:951984. doi: 10.3389/fimmu.2022.951984. eCollection 2022. Front Immunol. 2022. PMID: 35911774 Free PMC article.
References
-
- Hales LM, Knowles NJ, Reddy PS, et al. Complete genome sequence analysis of Seneca Valley virus-001, a novel oncolytic picornavirus. J Gen Virol. 2008;89(Pt 5):1265–1275. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
