Age-dependent expression changes of circadian system-related genes reveal a potentially conserved link to aging
- PMID: 34923482
- PMCID: PMC8751596
- DOI: 10.18632/aging.203788
Age-dependent expression changes of circadian system-related genes reveal a potentially conserved link to aging
Abstract
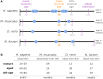
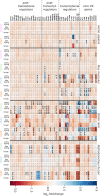
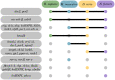
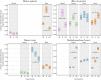
The circadian clock system influences the biology of life by establishing circadian rhythms in organisms, tissues, and cells, thus regulating essential biological processes based on the day/night cycle. Circadian rhythms change over a lifetime due to maturation and aging, and disturbances in the control of the circadian system are associated with several age-related pathologies. However, the impact of chronobiology and the circadian system on healthy organ and tissue aging remains largely unknown. Whether aging-related changes of the circadian system's regulation follow a conserved pattern across different species and tissues, hence representing a common driving force of aging, is unclear. Based on a cross-sectional transcriptome analysis covering 329 RNA-Seq libraries, we provide indications that the circadian system is subjected to aging-related gene alterations shared between evolutionarily distinct species, such as Homo sapiens, Mus musculus, Danio rerio, and Nothobranchius furzeri. We discovered differentially expressed genes by comparing tissue-specific transcriptional profiles of mature, aged, and old-age individuals and report on six genes (per2, dec2, cirp, klf10, nfil3, and dbp) of the circadian system, which show conserved aging-related expression patterns in four organs of the species examined. Our results illustrate how the circadian system and aging might influence each other in various tissues over a long lifespan and conceptually complement previous studies tracking short-term diurnal and nocturnal gene expression oscillations.
Keywords: RNA-Seq; aging; circadian clock system; circadian rhythm; inter-species comparison; longevity.
Conflict of interest statement
Figures

Similar articles
-
Defining the age-dependent and tissue-specific circadian transcriptome in male mice.Cell Rep. 2023 Jan 31;42(1):111982. doi: 10.1016/j.celrep.2022.111982. Epub 2023 Jan 9. Cell Rep. 2023. PMID: 36640301 Free PMC article.
-
Circadian regulation of metabolism and healthspan in Drosophila.Free Radic Biol Med. 2018 May 1;119:62-68. doi: 10.1016/j.freeradbiomed.2017.12.025. Epub 2017 Dec 19. Free Radic Biol Med. 2018. PMID: 29277395 Free PMC article. Review.
-
Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival.Sci Transl Med. 2016 Feb 3;8(324):324ra16. doi: 10.1126/scitranslmed.aad3305. Epub 2016 Feb 3. Sci Transl Med. 2016. PMID: 26843191 Free PMC article.
-
The intervertebral disc contains intrinsic circadian clocks that are regulated by age and cytokines and linked to degeneration.Ann Rheum Dis. 2017 Mar;76(3):576-584. doi: 10.1136/annrheumdis-2016-209428. Epub 2016 Aug 3. Ann Rheum Dis. 2017. PMID: 27489225 Free PMC article.
-
[Disrupted circadian rhythms and senescence].Nihon Rinsho. 2013 Dec;71(12):2091-6. Nihon Rinsho. 2013. PMID: 24437260 Review. Japanese.
Cited by
-
Age-dependent increase of cytoskeletal components in sensory axons in human skin.Front Cell Dev Biol. 2022 Nov 1;10:965382. doi: 10.3389/fcell.2022.965382. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36393849 Free PMC article.
-
Regulation of Circadian Genes Nr1d1 and Nr1d2 in Sex-Different Manners during Liver Aging.Int J Mol Sci. 2022 Sep 2;23(17):10032. doi: 10.3390/ijms231710032. Int J Mol Sci. 2022. PMID: 36077427 Free PMC article.
-
Age-Invariant Genes: Multi-Tissue Identification and Characterization of Murine Reference Genes.bioRxiv [Preprint]. 2024 Apr 13:2024.04.09.588721. doi: 10.1101/2024.04.09.588721. bioRxiv. 2024. PMID: 38645168 Free PMC article. Preprint.
-
Melatonin/Sericin Wound Healing Patches: Implications for Melanoma Therapy.Int J Mol Sci. 2024 Apr 29;25(9):4858. doi: 10.3390/ijms25094858. Int J Mol Sci. 2024. PMID: 38732075 Free PMC article. Review.
-
Current insights into transcriptional role(s) for the nutraceutical Withania somnifera in inflammation and aging.Front Nutr. 2024 May 3;11:1370951. doi: 10.3389/fnut.2024.1370951. eCollection 2024. Front Nutr. 2024. PMID: 38765810 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

