Cis-regulatory sequences in plants: Their importance, discovery, and future challenges
- PMID: 34918159
- PMCID: PMC8824567
- DOI: 10.1093/plcell/koab281
Cis-regulatory sequences in plants: Their importance, discovery, and future challenges
Abstract
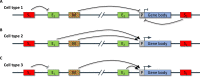
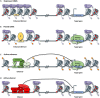
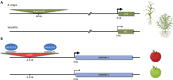
The identification and characterization of cis-regulatory DNA sequences and how they function to coordinate responses to developmental and environmental cues is of paramount importance to plant biology. Key to these regulatory processes are cis-regulatory modules (CRMs), which include enhancers and silencers. Despite the extraordinary advances in high-quality sequence assemblies and genome annotations, the identification and understanding of CRMs, and how they regulate gene expression, lag significantly behind. This is especially true for their distinguishing characteristics and activity states. Here, we review the current knowledge on CRMs and breakthrough technologies enabling identification, characterization, and validation of CRMs; we compare the genomic distributions of CRMs with respect to their target genes between different plant species, and discuss the role of transposable elements harboring CRMs in the evolution of gene expression. This is an exciting time to study cis-regulomes in plants; however, significant existing challenges need to be overcome to fully understand and appreciate the role of CRMs in plant biology and in crop improvement.
© The Author(s) 2021. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures




Similar articles
-
The prevalence, evolution and chromatin signatures of plant regulatory elements.Nat Plants. 2019 Dec;5(12):1250-1259. doi: 10.1038/s41477-019-0548-z. Epub 2019 Nov 18. Nat Plants. 2019. PMID: 31740772
-
De novo prediction of cis-regulatory elements and modules through integrative analysis of a large number of ChIP datasets.BMC Genomics. 2014 Dec 2;15:1047. doi: 10.1186/1471-2164-15-1047. BMC Genomics. 2014. PMID: 25442502 Free PMC article.
-
Identifying transcriptional cis-regulatory modules in animal genomes.Wiley Interdiscip Rev Dev Biol. 2015 Mar-Apr;4(2):59-84. doi: 10.1002/wdev.168. Epub 2014 Dec 29. Wiley Interdiscip Rev Dev Biol. 2015. PMID: 25704908 Free PMC article. Review.
-
Plant Enhancers: A Call for Discovery.Trends Plant Sci. 2016 Nov;21(11):974-987. doi: 10.1016/j.tplants.2016.07.013. Epub 2016 Sep 2. Trends Plant Sci. 2016. PMID: 27593567 Review.
-
Predicting tissue specific cis-regulatory modules in the human genome using pairs of co-occurring motifs.BMC Bioinformatics. 2012 Feb 7;13:25. doi: 10.1186/1471-2105-13-25. BMC Bioinformatics. 2012. PMID: 22313678 Free PMC article.
Cited by
-
iCREPCP: A deep learning-based web server for identifying base-resolution cis-regulatory elements within plant core promoters.Plant Commun. 2023 Jan 9;4(1):100455. doi: 10.1016/j.xplc.2022.100455. Epub 2022 Sep 28. Plant Commun. 2023. PMID: 36171722 Free PMC article. No abstract available.
-
Genome-Wide Identification and Characterization of the Polyamine Uptake Transporter (Put) Gene Family in Tomatoes and the Role of Put2 in Response to Salt Stress.Antioxidants (Basel). 2023 Jan 18;12(2):228. doi: 10.3390/antiox12020228. Antioxidants (Basel). 2023. PMID: 36829787 Free PMC article.
-
Genome wide identification and evolutionary analysis of vat like NBS-LRR genes potentially associated with resistance to aphids in cotton.Genetica. 2023 Apr;151(2):119-131. doi: 10.1007/s10709-023-00181-1. Epub 2023 Jan 30. Genetica. 2023. PMID: 36717534
-
Genome-Wide Survey and Expression Analyses of Hexokinase Family in Poplar (Populus trichocarpa).Plants (Basel). 2022 Aug 3;11(15):2025. doi: 10.3390/plants11152025. Plants (Basel). 2022. PMID: 35956502 Free PMC article.
-
Identification of a Unique Genomic Region in Sweet Chestnut (Castanea sativa Mill.) That Controls Resistance to Asian Chestnut Gall Wasp Dryocosmus kuriphilus Yasumatsu.Plants (Basel). 2024 May 14;13(10):1355. doi: 10.3390/plants13101355. Plants (Basel). 2024. PMID: 38794426 Free PMC article.
References
-
- Andersson R, Refsing Andersen P, Valen E, Core LJ, Bornholdt J, Boyd M, Heick Jensen T, Sandelin A (2014b) Nuclear stability and transcriptional directionality separate functionally distinct RNA species. Nat Commun 5: 5336. - PubMed
-
- Andersson R, Sandelin A (2020) Determinants of enhancer and promoter activities of regulatory elements. Nat Rev Genet 21: 71–87 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

