TRPM7 N-terminal region forms complexes with calcium binding proteins CaM and S100A1
- PMID: 34917797
- PMCID: PMC8645431
- DOI: 10.1016/j.heliyon.2021.e08490
TRPM7 N-terminal region forms complexes with calcium binding proteins CaM and S100A1
Abstract
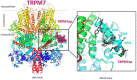
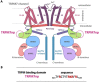
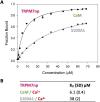
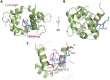
Transient receptor potential melastatin 7 (TRPM7) represents melastatin TRP channel with two significant functions, cation permeability and kinase activity. TRPM7 is widely expressed among tissues and is therefore involved in a variety of cellular functions representing mainly Mg2+ homeostasis, cellular Ca2+ flickering, and the regulation of DNA transcription by a cleaved kinase domain translocated to the nucleus. TRPM7 participates in several important biological processes in the nervous and cardiovascular systems. Together with the necessary function of the TRPM7 in these tissues and its recently analyzed overall structure, this channel requires further studies leading to the development of potential therapeutic targets. Here we present the first study investigating the N-termini of TRPM7 with binding regions for important intracellular modulators calmodulin (CaM) and calcium-binding protein S1 (S100A1) using in vitro and in silico approaches. Molecular simulations of the discovered complexes reveal their potential binding interfaces with common interaction patterns and the important role of basic residues present in the N-terminal binding region of TRPM.
Keywords: Binding region; CaM; Calcium; Fluorescence anisotropy; S100A1; TRPM7.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
TRPM6 N-Terminal CaM- and S100A1-Binding Domains.Int J Mol Sci. 2019 Sep 9;20(18):4430. doi: 10.3390/ijms20184430. Int J Mol Sci. 2019. PMID: 31505788 Free PMC article.
-
Mapping of CaM, S100A1 and PIP2-Binding Epitopes in the Intracellular N- and C-Termini of TRPM4.Int J Mol Sci. 2020 Jun 17;21(12):4323. doi: 10.3390/ijms21124323. Int J Mol Sci. 2020. PMID: 32560560 Free PMC article.
-
TRPM5 Channel Binds Calcium-Binding Proteins Calmodulin and S100A1.Biochemistry. 2022 Mar 15;61(6):413-423. doi: 10.1021/acs.biochem.1c00647. Epub 2022 Feb 28. Biochemistry. 2022. PMID: 35225608
-
The TRPM7 Channel in the Nervous and Cardiovascular Systems.Curr Protein Pept Sci. 2020;21(10):985-992. doi: 10.2174/1389203721666200605170938. Curr Protein Pept Sci. 2020. PMID: 32503408 Review.
-
Waixenicin A, a marine-derived TRPM7 inhibitor: a promising CNS drug lead.Acta Pharmacol Sin. 2020 Dec;41(12):1519-1524. doi: 10.1038/s41401-020-00512-4. Epub 2020 Sep 29. Acta Pharmacol Sin. 2020. PMID: 32994545 Free PMC article. Review.
Cited by
-
Interaction of Calmodulin with TRPM: An Initiator of Channel Modulation.Int J Mol Sci. 2023 Oct 13;24(20):15162. doi: 10.3390/ijms242015162. Int J Mol Sci. 2023. PMID: 37894842 Free PMC article. Review.
-
Effect of truncation on TRPM7 channel activity.Channels (Austin). 2023 Dec;17(1):2200874. doi: 10.1080/19336950.2023.2200874. Channels (Austin). 2023. PMID: 37040321 Free PMC article.
References
-
- Ryazanova L.V., Dorovkov M.V., Ansari A., Ryazanov A.G. Characterization of the protein kinase activity of TRPM7/ChaK1, a protein kinase fused to the transient receptor potential ion channel. J. Biol. Chem. 2004;279(5):3708–3716. - PubMed
-
- Kim T.Y., Shin S.K., Song M.-Y., Lee J.E., Park K.-S. Identification of the phosphorylation sites on intact TRPM7 channels from mammalian cells. Biochem. Biophys. Res. Commun. 2012;417(3):1030–1034. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

