Genome-Wide Characterization of Zebrafish Endogenous Retroviruses Reveals Unexpected Diversity in Genetic Organizations and Functional Potentials
- PMID: 34908463
- PMCID: PMC8672886
- DOI: 10.1128/spectrum.02254-21
Genome-Wide Characterization of Zebrafish Endogenous Retroviruses Reveals Unexpected Diversity in Genetic Organizations and Functional Potentials
Abstract
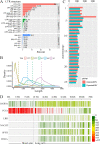
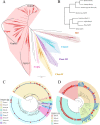
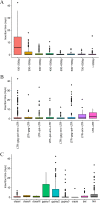
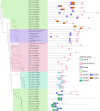
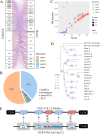
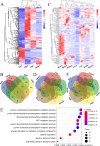
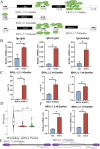
Endogenous retroviruses (ERVs) occupy a substantial fraction of mammalian genomes. However, whether ERVs extensively exist in ancient vertebrates remains unexplored. Here, we performed a genome-wide characterization of ERVs in a zebrafish (Danio rerio) model. Approximately 3,315 ERV-like elements (DrERVs) were identified as Gypsy, Copia, Bel, and class I-III groups. DrERVs accounted for approximately 2.3% of zebrafish genome and were distributed in all 25 chromosomes, with a remarkable bias on chromosome 4. Gypsy and class I are the two most abundant groups with earlier insertion times. The vast majority of the DrERVs have varied structural defects. A total of 509 gag and 71 env genes with coding potentials were detected. The env-coding elements were well-characterized and classified into four subgroups. A ERV-E4.8.43-DanRer element shows high similarity with HERV9NC-int in humans and analogous sequences were detected in species spanning from fish to mammals. RNA-seq data showed that hundreds of DrERVs were expressed in embryos and tissues under physiological conditions, and most of them exhibited stage and tissue specificity. Additionally, 421 DrERVs showed strong responsiveness to virus infection. A unique group of DrERVs with immune-relevant genes, such as fga, ddx41, ftr35, igl1c3, and tbk1, instead of intrinsic viral genes were identified. These DrERVs are regulated by transcriptional factors binding at the long terminal repeats. This study provided a survey of the composition, phylogeny, and potential functions of ERVs in a fish model, which benefits the understanding of the evolutionary history of ERVs from fish to mammals. IMPORTANCE Endogenous retroviruses (ERVs) are relics of past infection that constitute up to 8% of the human genome. Understanding the genetic evolution of the ERV family and the interplay of ERVs and encoded RNAs and proteins with host function has become a new frontier in biology. Fish, as the most primitive vertebrate host for retroviruses, is an indispensable integral part for such investigations. In the present study, we report the genome-wide characterization of ERVs in zebrafish, an attractive model organism of ancient vertebrates from multiple perspectives, including composition, genomic organization, chromosome distribution, classification, phylogeny, insertion time, characterization of gag and env genes, and expression profiles in embryos and tissues. The result helps uncover the evolutionarily conserved and fish-specific ERVs, as well as the immune-relevant ERVs in response to virus infection. This study demonstrates the previously unrecognized abundance, diversification, and extensive activity of ERVs at the early stage of ERV evolution.
Keywords: endogenous retrovirus; evolution; expression; structure; zebrafish.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
A novel class III endogenous retrovirus with a class I envelope gene in African frogs with an intact genome and developmentally regulated transcripts in Xenopus tropicalis.Retrovirology. 2021 Jul 14;18(1):20. doi: 10.1186/s12977-021-00564-2. Retrovirology. 2021. PMID: 34261506 Free PMC article.
-
Distribution, Diversity, and Evolution of Endogenous Retroviruses in Perissodactyl Genomes.J Virol. 2018 Nov 12;92(23):e00927-18. doi: 10.1128/JVI.00927-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30209175 Free PMC article.
-
Identification of a group of Mus dunni endogenous virus-like endogenous retroviruses from the C57BL/6J mouse genome: proviral genomes, strain distribution, expression characteristics, and genomic integration profile.Chromosome Res. 2012 Oct;20(7):859-74. doi: 10.1007/s10577-012-9322-z. Epub 2012 Nov 30. Chromosome Res. 2012. PMID: 23197326 Free PMC article.
-
Origins and evolutionary consequences of ancient endogenous retroviruses.Nat Rev Microbiol. 2019 Jun;17(6):355-370. doi: 10.1038/s41579-019-0189-2. Nat Rev Microbiol. 2019. PMID: 30962577 Review.
-
The evolution, distribution and diversity of endogenous retroviruses.Virus Genes. 2003 May;26(3):291-315. doi: 10.1023/a:1024455415443. Virus Genes. 2003. PMID: 12876457 Review.
Cited by
-
Microbe transmission from pet shop to lab-reared zebrafish reveals a pathogenic birnavirus.PLoS Biol. 2024 May 30;22(5):e3002606. doi: 10.1371/journal.pbio.3002606. eCollection 2024 May. PLoS Biol. 2024. PMID: 38814944 Free PMC article.
-
Viruses in astrobiology.Front Microbiol. 2022 Oct 26;13:1032918. doi: 10.3389/fmicb.2022.1032918. eCollection 2022. Front Microbiol. 2022. PMID: 36386652 Free PMC article. Review.
-
Essential role of an ERV-derived Env38 protein in adaptive humoral immunity against an exogenous SVCV infection in a zebrafish model.PLoS Pathog. 2023 Apr 4;19(4):e1011222. doi: 10.1371/journal.ppat.1011222. eCollection 2023 Apr. PLoS Pathog. 2023. PMID: 37014912 Free PMC article.
-
Comprehensive Transcriptome Analysis Reveals Sex-Specific Alternative Splicing Events in Zebrafish Gonads.Life (Basel). 2022 Sep 16;12(9):1441. doi: 10.3390/life12091441. Life (Basel). 2022. PMID: 36143477 Free PMC article.
-
Unique Structure and Distinctive Properties of the Ancient and Ubiquitous Gamma-Type Envelope Glycoprotein.Viruses. 2023 Jan 18;15(2):274. doi: 10.3390/v15020274. Viruses. 2023. PMID: 36851488 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

