Cyclin/Forkhead-mediated coordination of cyclin waves: an autonomous oscillator rationalizing the quantitative model of Cdk control for budding yeast
- PMID: 34903735
- PMCID: PMC8668886
- DOI: 10.1038/s41540-021-00201-w
Cyclin/Forkhead-mediated coordination of cyclin waves: an autonomous oscillator rationalizing the quantitative model of Cdk control for budding yeast
Abstract
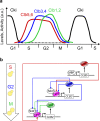
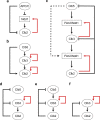
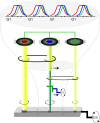
Networks of interacting molecules organize topology, amount, and timing of biological functions. Systems biology concepts required to pin down 'network motifs' or 'design principles' for time-dependent processes have been developed for the cell division cycle, through integration of predictive computer modeling with quantitative experimentation. A dynamic coordination of sequential waves of cyclin-dependent kinases (cyclin/Cdk) with the transcription factors network offers insights to investigate how incompatible processes are kept separate in time during the eukaryotic cell cycle. Here this coordination is discussed for the Forkhead transcription factors in light of missing gaps in the current knowledge of cell cycle control in budding yeast. An emergent design principle is proposed where cyclin waves are synchronized by a cyclin/Cdk-mediated feed-forward regulation through the Forkhead as a transcriptional timer. This design is rationalized by the bidirectional interaction between mitotic cyclins and the Forkhead transcriptional timer, resulting in an autonomous oscillator that may be instrumental for a well-timed progression throughout the cell cycle. The regulation centered around the cyclin/Cdk-Forkhead axis can be pivotal to timely coordinate cell cycle dynamics, thereby to actuate the quantitative model of Cdk control.
© 2021. The Author(s).
Conflict of interest statement
The author declares no competing interests.
Figures
Similar articles
-
Quantitative model of eukaryotic Cdk control through the Forkhead CONTROLLER.NPJ Syst Biol Appl. 2021 Jun 11;7(1):28. doi: 10.1038/s41540-021-00187-5. NPJ Syst Biol Appl. 2021. PMID: 34117265 Free PMC article. Review.
-
Clb3-centered regulations are recurrent across distinct parameter regions in minimal autonomous cell cycle oscillator designs.NPJ Syst Biol Appl. 2020 Apr 3;6(1):8. doi: 10.1038/s41540-020-0125-0. NPJ Syst Biol Appl. 2020. PMID: 32245958 Free PMC article.
-
The cyclin family of budding yeast: abundant use of a good idea.Trends Genet. 1998 Feb;14(2):66-72. doi: 10.1016/s0168-9525(97)01322-x. Trends Genet. 1998. PMID: 9520600 Review.
-
Global control of cell-cycle transcription by coupled CDK and network oscillators.Nature. 2008 Jun 12;453(7197):944-7. doi: 10.1038/nature06955. Epub 2008 May 7. Nature. 2008. PMID: 18463633 Free PMC article.
-
Periodic cyclin-Cdk activity entrains an autonomous Cdc14 release oscillator.Cell. 2010 Apr 16;141(2):268-79. doi: 10.1016/j.cell.2010.03.021. Cell. 2010. PMID: 20403323 Free PMC article.
Cited by
-
A yeast cell cycle model integrating stress, signaling, and physiology.FEMS Yeast Res. 2022 Jun 30;22(1):foac026. doi: 10.1093/femsyr/foac026. FEMS Yeast Res. 2022. PMID: 35617157 Free PMC article.
-
Unveiling Forkhead-mediated regulation of yeast cell cycle and metabolic networks.Comput Struct Biotechnol J. 2022 Apr 7;20:1743-1751. doi: 10.1016/j.csbj.2022.03.033. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35495119 Free PMC article.
References
-
- Nasmyth K. Control of the yeast cell cycle by the Cdc28 protein kinase. Curr. Opin. Cell Biol. 1993;5:166–179. - PubMed
-
- Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405–412. - PubMed
-
- Futcher B. Cyclins and the wiring of the yeast cell cycle. Yeast. 1996;12:1635–1646. - PubMed
-
- Andrews B, Measday V. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. - PubMed
-
- Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nat. Rev. Mol. Cell Biol. 2007;8:149–160. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

