M-MuLV reverse transcriptase: Selected properties and improved mutants
- PMID: 34900141
- PMCID: PMC8640165
- DOI: 10.1016/j.csbj.2021.11.030
M-MuLV reverse transcriptase: Selected properties and improved mutants
Abstract
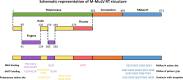
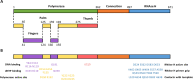
Reverse transcriptases (RTs) are enzymes synthesizing DNA using RNA as the template and serving as the standard tools in modern biotechnology and molecular diagnostics. To date, the most commonly used reverse transcriptase is the enzyme from Moloney murine leukemia virus, M-MuLV RT. Since its discovery, M-MuLV RT has become indispensable for modern RNA studies; the range of M-MuLV RT applications is vast, from scientific tasks to clinical testing of human pathogens. This review will give a brief description of the structure, thermal stability, processivity, and fidelity, focusing on improving M-MuLV RT for practical usage.
Keywords: Fusion proteins; M-MuLV RT; Random mutagenesis; Reverse transcriptase; Site-directed mutagenesis.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Comparison of Reverse Transcriptase (RT) Activities of Various M-MuLV RTs for RT-LAMP Assays.Biology (Basel). 2022 Dec 13;11(12):1809. doi: 10.3390/biology11121809. Biology (Basel). 2022. PMID: 36552320 Free PMC article.
-
The attachment of a DNA-binding Sso7d-like protein improves processivity and resistance to inhibitors of M-MuLV reverse transcriptase.FEBS Lett. 2020 Dec;594(24):4338-4356. doi: 10.1002/1873-3468.13934. Epub 2020 Oct 5. FEBS Lett. 2020. PMID: 32970841
-
Mechanistic differences in RNA-dependent DNA polymerization and fidelity between murine leukemia virus and HIV-1 reverse transcriptases.J Biol Chem. 2005 Apr 1;280(13):12190-200. doi: 10.1074/jbc.M412859200. Epub 2005 Jan 10. J Biol Chem. 2005. PMID: 15644314 Free PMC article.
-
Lysine 152 of MuLV reverse transcriptase is required for the integrity of the active site.Biochemistry. 2002 Dec 17;41(50):14831-42. doi: 10.1021/bi0258389. Biochemistry. 2002. PMID: 12475231
-
Viral reverse transcriptases.Virus Res. 2017 Apr 15;234:153-176. doi: 10.1016/j.virusres.2016.12.019. Epub 2016 Dec 30. Virus Res. 2017. PMID: 28043823 Review.
Cited by
-
Reverse transcription of plasma-derived HIV-1 RNA generates multiple artifacts through tRNA(Lys-3)-priming.Microbiol Spectr. 2024 Apr 2;12(4):e0387223. doi: 10.1128/spectrum.03872-23. Epub 2024 Mar 5. Microbiol Spectr. 2024. PMID: 38442427 Free PMC article.
-
Profiling of RNA-binding protein binding sites by in situ reverse transcription-based sequencing.Nat Methods. 2024 Feb;21(2):247-258. doi: 10.1038/s41592-023-02146-w. Epub 2024 Jan 10. Nat Methods. 2024. PMID: 38200227 Free PMC article.
-
Prime editing: Mechanism insight and recent applications in plants.Plant Biotechnol J. 2024 Jan;22(1):19-36. doi: 10.1111/pbi.14188. Epub 2023 Oct 4. Plant Biotechnol J. 2024. PMID: 37794706 Free PMC article. Review.
-
CleanUpRNAseq: An R/Bioconductor Package for Detecting and Correcting DNA Contamination in RNA-Seq Data.BioTech (Basel). 2024 Aug 3;13(3):30. doi: 10.3390/biotech13030030. BioTech (Basel). 2024. PMID: 39189209 Free PMC article.
-
Structural basis for pegRNA-guided reverse transcription by a prime editor.Nature. 2024 Jul;631(8019):224-231. doi: 10.1038/s41586-024-07497-8. Epub 2024 May 29. Nature. 2024. PMID: 38811740 Free PMC article.
References
-
- Gross L. “Spontaneous” leukemia developing in C3H mice following inoculation in infancy, with AK-leukemic extracts, or AK-embrvos. Proc Soc Exp Biol Med. 1951;76:27–32. - PubMed
-
- Gross L. Filterable agent causing leukemia following inoculation into newborn. Tex Rep Biol Med. 1957;15 - PubMed
-
- Moloney J.B. Properties of a leukemia virus. Natl Cancer Inst Monogr. 1960;4:7–37. - PubMed
-
- Moloney J.B. Biological studies on a lymphoid-leukemia virus extracted from sarcoma 37. I. Origin and introductory investigations. J Natl Cancer Inst. 1960;24:933–951. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources