Energetics of lipid transport by the ABC transporter MsbA is lipid dependent
- PMID: 34887543
- PMCID: PMC8660845
- DOI: 10.1038/s42003-021-02902-8
Energetics of lipid transport by the ABC transporter MsbA is lipid dependent
Abstract
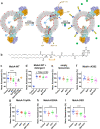
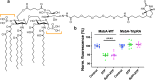
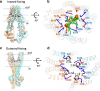
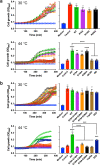
The ABC multidrug exporter MsbA mediates the translocation of lipopolysaccharides and phospholipids across the plasma membrane in Gram-negative bacteria. Although MsbA is structurally well characterised, the energetic requirements of lipid transport remain unknown. Here, we report that, similar to the transport of small-molecule antibiotics and cytotoxic agents, the flopping of physiologically relevant long-acyl-chain 1,2-dioleoyl (C18)-phosphatidylethanolamine in proteoliposomes requires the simultaneous input of ATP binding and hydrolysis and the chemical proton gradient as sources of metabolic energy. In contrast, the flopping of the large hexa-acylated (C12-C14) Lipid-A anchor of lipopolysaccharides is only ATP dependent. This study demonstrates that the energetics of lipid transport by MsbA is lipid dependent. As our mutational analyses indicate lipid and drug transport via the central binding chamber in MsbA, the lipid availability in the membrane can affect the drug transport activity and vice versa.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Drug-lipid A interactions on the Escherichia coli ABC transporter MsbA.J Bacteriol. 2005 Sep;187(18):6363-9. doi: 10.1128/JB.187.18.6363-6369.2005. J Bacteriol. 2005. PMID: 16159769 Free PMC article.
-
The ATP binding cassette multidrug transporter LmrA and lipid transporter MsbA have overlapping substrate specificities.J Biol Chem. 2003 Sep 12;278(37):35193-8. doi: 10.1074/jbc.M306226200. Epub 2003 Jul 2. J Biol Chem. 2003. PMID: 12842882
-
Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis.J Biol Chem. 1998 May 15;273(20):12466-75. doi: 10.1074/jbc.273.20.12466. J Biol Chem. 1998. PMID: 9575204
-
Multidrug transporters and antibiotic resistance in Lactococcus lactis.Biochim Biophys Acta. 2002 Sep 10;1555(1-3):1-7. doi: 10.1016/s0005-2728(02)00246-3. Biochim Biophys Acta. 2002. PMID: 12206883 Review.
-
MsbA: an ABC transporter paradigm.Biochem Soc Trans. 2021 Dec 17;49(6):2917-2927. doi: 10.1042/BST20211030. Biochem Soc Trans. 2021. PMID: 34821931 Review.
Cited by
-
Reconstitution of ATP-dependent lipid transporters: gaining insight into molecular characteristics, regulation, and mechanisms.Biosci Rep. 2023 Aug 31;43(8):BSR20221268. doi: 10.1042/BSR20221268. Biosci Rep. 2023. PMID: 37417269 Free PMC article. Review.
-
Drug-dependent inhibition of nucleotide hydrolysis in the heterodimeric ABC multidrug transporter PatAB from Streptococcus pneumoniae.FEBS J. 2022 Jul;289(13):3770-3788. doi: 10.1111/febs.16366. Epub 2022 Feb 11. FEBS J. 2022. PMID: 35066976 Free PMC article.
-
Double and triple thermodynamic mutant cycles reveal the basis for specific MsbA-lipid interactions.bioRxiv [Preprint]. 2023 Nov 6:2023.07.03.547565. doi: 10.1101/2023.07.03.547565. bioRxiv. 2023. Update in: Elife. 2024 Jan 22;12:RP91094. doi: 10.7554/eLife.91094 PMID: 37461710 Free PMC article. Updated. Preprint.
-
Changes in the Human Gut Microbiome Caused by the Short-Term Impact of Lactic Acid Bacteria Consumption in Healthy People.Probiotics Antimicrob Proteins. 2024 Aug;16(4):1240-1250. doi: 10.1007/s12602-023-10111-4. Epub 2023 Jun 26. Probiotics Antimicrob Proteins. 2024. PMID: 37365419
-
Cerastecins inhibit membrane lipooligosaccharide transport in drug-resistant Acinetobacter baumannii.Nat Microbiol. 2024 May;9(5):1244-1255. doi: 10.1038/s41564-024-01667-0. Epub 2024 Apr 22. Nat Microbiol. 2024. PMID: 38649414
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

