The Emerging Role of Non-Coding RNAs in Pituitary Gland Tumors and Meningioma
- PMID: 34885097
- PMCID: PMC8656547
- DOI: 10.3390/cancers13235987
The Emerging Role of Non-Coding RNAs in Pituitary Gland Tumors and Meningioma
Abstract
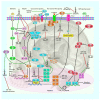
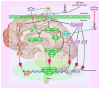
Long non-coding RNAs (lncRNAs), microRNAs (miRNAs), and circular RNAs (circRNAs) are non-coding transcripts which are involved in the pathogenesis of pituitary gland tumors. LncRNAs that participate in the pathogenesis of pituitary gland tumors mainly serve as sponges for miRNAs. CLRN1-AS1/miR-217, XIST/miR-424-5p, H19/miR-93a, LINC00473/miR-502-3p, SNHG7/miR-449a, MEG8/miR-454-3p, MEG3/miR-23b-3p, MEG3/miR-376B-3P, SNHG6/miR-944, PCAT6/miR-139-3p, lncRNA-m433s1/miR-433, TUG1/miR-187-3p, SNHG1/miR-187-3p, SNHG1/miR-302, SNHG1/miR-372, SNHG1/miR-373, and SNHG1/miR-520 are identified lncRNA/miRNA pairs that are involved in this process. Hsa_circ_0001368 and circOMA1 are two examples of circRNAs that contribute to the pathogenesis of pituitary gland tumors. Meanwhile, SNHG1, LINC00702, LINC00460, and MEG3 have been found to partake in the pathogenesis of meningioma. In the current review, we describe the role of non-coding RNAs in two types of brain tumors, i.e., pituitary tumors and meningioma.
Keywords: circRNA; lncRNA; meningioma; miRNA; pituitary gland cancer.
Conflict of interest statement
The authors declare they have no conflicts of interests.
Figures
Similar articles
-
Interplay between lncRNA/miRNA and Wnt/ß-catenin signaling in brain cancer tumorigenesis.EXCLI J. 2023 Nov 28;22:1211-1222. doi: 10.17179/excli2023-6490. eCollection 2023. EXCLI J. 2023. PMID: 38204968 Free PMC article. Review.
-
The key roles of non-coding RNAs in the pathophysiology of hypertension.Eur J Pharmacol. 2022 Sep 15;931:175220. doi: 10.1016/j.ejphar.2022.175220. Epub 2022 Aug 19. Eur J Pharmacol. 2022. PMID: 35995213 Review.
-
Regulatory Role of Non-Coding RNAs on Immune Responses During Sepsis.Front Immunol. 2021 Dec 9;12:798713. doi: 10.3389/fimmu.2021.798713. eCollection 2021. Front Immunol. 2021. PMID: 34956235 Free PMC article. Review.
-
Non-coding RNAs as Genetic Biomarkers for the Diagnosis, Prognosis, Radiosensitivity, and Histopathologic Grade of Meningioma.Cureus. 2023 Feb 3;15(2):e34593. doi: 10.7759/cureus.34593. eCollection 2023 Feb. Cureus. 2023. PMID: 36883085 Free PMC article. Review.
-
SNHG1 promotes malignant biological behaviors of glioma cells via microRNA-154-5p/miR-376b-3p- FOXP2- KDM5B participating positive feedback loop.J Exp Clin Cancer Res. 2019 Feb 6;38(1):59. doi: 10.1186/s13046-019-1063-9. J Exp Clin Cancer Res. 2019. PMID: 30728054 Free PMC article.
Cited by
-
Circulating Noncoding RNAs in Pituitary Neuroendocrine Tumors-Two Sides of the Same Coin.Int J Mol Sci. 2022 May 4;23(9):5122. doi: 10.3390/ijms23095122. Int J Mol Sci. 2022. PMID: 35563510 Free PMC article. Review.
-
Expression of cAMP and oxidative phosphorylation-related lncRNAs in non-functioning pituitary adenomas.J Cell Mol Med. 2023 Dec;27(24):4195-4201. doi: 10.1111/jcmm.18011. Epub 2023 Nov 6. J Cell Mol Med. 2023. PMID: 37933082 Free PMC article.
-
The hallmarks of cancer… in pituitary tumors?Rev Endocr Metab Disord. 2023 Apr;24(2):177-190. doi: 10.1007/s11154-022-09777-y. Epub 2022 Dec 31. Rev Endocr Metab Disord. 2023. PMID: 36586070 Review.
-
Role of MicroRNA-502-3p in Human Diseases.Pharmaceuticals (Basel). 2023 Apr 2;16(4):532. doi: 10.3390/ph16040532. Pharmaceuticals (Basel). 2023. PMID: 37111289 Free PMC article. Review.
-
Long Noncoding RNAs Expressed in Mouse Pituitary Development and Mature Hormone-Producing Cells.Endocrinology. 2024 Oct 30;165(12):bqae147. doi: 10.1210/endocr/bqae147. Endocrinology. 2024. PMID: 39487735
References
-
- Grillone K., Riillo C., Scionti F., Rocca R., Tradigo G., Guzzi P.H., Alcaro S., Di Martino M.T., Tagliaferri P., Tassone P. Non-coding RNAs in cancer: Platforms and strategies for investigating the genomic “dark matter”. J. Exp. Clin. Cancer Res. 2020;39:1–19. doi: 10.1186/s13046-020-01622-x. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

