Conformational Tuning of Amylin by Charged Styrene-Maleic-Acid Copolymers
- PMID: 34883118
- PMCID: PMC8752516
- DOI: 10.1016/j.jmb.2021.167385
Conformational Tuning of Amylin by Charged Styrene-Maleic-Acid Copolymers
Abstract
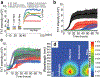
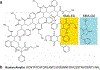
Human amylin forms structurally heterogeneous amyloids that have been linked to type-2 diabetes. Thus, understanding the molecular interactions governing amylin aggregation can provide mechanistic insights in its pathogenic formation. Here, we demonstrate that fibril formation of amylin is altered by synthetic amphipathic copolymer derivatives of the styrene-maleic-acid (SMAQA and SMAEA). High-speed AFM is used to follow the real-time aggregation of amylin by observing the rapid formation of de novo globular oligomers and arrestment of fibrillation by the positively-charged SMAQA. We also observed an accelerated fibril formation in the presence of the negatively-charged SMAEA. These findings were further validated by fluorescence, SOFAST-HMQC, DOSY and STD NMR experiments. Conformational analysis by CD and FT-IR revealed that the SMA copolymers modulate the conformation of amylin aggregates. While the species formed with SMAQA are α-helical, the ones formed with SMAEA are rich in β-sheet structure. The interacting interfaces between SMAEA or SMAQA and amylin are mapped by NMR and microseconds all-atom MD simulation. SMAEA displayed π-π interaction with Phe23, electrostatic π-cation interaction with His18 and hydrophobic packing with Ala13 and Val17; whereas SMAQA showed a selective interaction with amylin's C terminus (residues 31-37) that belongs to one of the two β-sheet regions (residues 14-19 and 31-36) involved in amylin fibrillation. Toxicity analysis showed both SMA copolymers to be non-toxic in vitro and the amylin species formed with the copolymers showed minimal deformity to zebrafish embryos. Together, this study demonstrates that chemical tools, such as copolymers, can be used to modulate amylin aggregation, alter the conformation of species.
Keywords: IAPP; SMA copolymer; amylin; amyloid; type-II diabetes.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The polymers used in this study SMAEA and SMAQA are produced in the Ramamoorthy lab at Michigan. They are US patented.
Figures





Similar articles
-
Full length amylin oligomer aggregation: insights from molecular dynamics simulations and implications for design of aggregation inhibitors.J Biomol Struct Dyn. 2014;32(10):1651-69. doi: 10.1080/07391102.2013.832635. Epub 2013 Sep 13. J Biomol Struct Dyn. 2014. PMID: 24028418
-
Rapid assessment of human amylin aggregation and its inhibition by copper(II) ions by laser ablation electrospray ionization mass spectrometry with ion mobility separation.Anal Chem. 2015 Oct 6;87(19):9829-9837. doi: 10.1021/acs.analchem.5b02217. Anal Chem. 2015. PMID: 26352401 Free PMC article.
-
Fractal self-assembly and aggregation of human amylin.Soft Matter. 2020 Mar 28;16(12):3143-3153. doi: 10.1039/c9sm02463h. Epub 2020 Mar 11. Soft Matter. 2020. PMID: 32159545
-
Human Amylin: From Pathology to Physiology and Pharmacology.Curr Protein Pept Sci. 2019;20(9):944-957. doi: 10.2174/1389203720666190328111833. Curr Protein Pept Sci. 2019. PMID: 30919775 Review.
-
Role of mass spectrometry in the study of interactions between amylin and metal ions.Mass Spectrom Rev. 2023 May;42(3):984-1007. doi: 10.1002/mas.21732. Epub 2021 Sep 24. Mass Spectrom Rev. 2023. PMID: 34558100 Review.
Cited by
-
Amyloid Oligomers: A Joint Experimental/Computational Perspective on Alzheimer's Disease, Parkinson's Disease, Type II Diabetes, and Amyotrophic Lateral Sclerosis.Chem Rev. 2021 Feb 24;121(4):2545-2647. doi: 10.1021/acs.chemrev.0c01122. Epub 2021 Feb 5. Chem Rev. 2021. PMID: 33543942 Free PMC article. Review.
-
Single-Molecule Kinetic Observation of Antibody Interactions with Growing Amyloid β Fibrils.J Am Chem Soc. 2024 Nov 20;146(46):31518-31528. doi: 10.1021/jacs.4c08841. Epub 2024 Oct 24. J Am Chem Soc. 2024. PMID: 39445702 Free PMC article.
-
On-Pathway Oligomer of Human Islet Amyloid Polypeptide Induced and Stabilized by Mechanical Rotation During MAS NMR.bioRxiv [Preprint]. 2023 Jul 8:2023.07.06.547982. doi: 10.1101/2023.07.06.547982. bioRxiv. 2023. Update in: J Phys Chem Lett. 2023 Aug 31;14(34):7644-7649. doi: 10.1021/acs.jpclett.3c02009. PMID: 37461639 Free PMC article. Updated. Preprint.
-
Spatiotemporal resolution in high-speed atomic force microscopy for studying biological macromolecules in action.Microscopy (Oxf). 2023 Apr 6;72(2):151-161. doi: 10.1093/jmicro/dfad011. Microscopy (Oxf). 2023. PMID: 36744614 Free PMC article. Review.
-
On-Pathway Oligomer of Human Islet Amyloid Polypeptide Induced and Stabilized by Mechanical Rotation during Magic Angle Spinning Nuclear Magnetic Resonance.J Phys Chem Lett. 2023 Aug 31;14(34):7644-7649. doi: 10.1021/acs.jpclett.3c02009. Epub 2023 Aug 21. J Phys Chem Lett. 2023. PMID: 37602799 Free PMC article.
References
-
- Gurlo T, Ryazantsev S, Huang CJ, Yeh MW, Reber HA, Hines OJ, O’Brien TD, Glabe CG, Butler PC, Evidence for proteotoxicity in β cells in type 2 diabetes: Toxic islet amyloid polypeptide oligomers form intracellularly in the secretory pathway, American Journal of Pathology. 176 (2010) 861–869. 10.2353/ajpath.2010.090532. - DOI - PMC - PubMed
-
- Lin CY, Gurlo T, Kayed R, Butler AE, Haataja L, Glabe CG, Butler PC, Toxic human islet amyloid polypeptide (h-IAPP) oligomers are intracellular, and vaccination to induce anti-toxic oligomer antibodies does not prevent h-IAPP-induced β-cell apoptosis in h-IAPP transgenic mice, Diabetes. 56 (2007) 1324–1332. 10.2337/db06-1579. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

