OME-NGFF: a next-generation file format for expanding bioimaging data-access strategies
- PMID: 34845388
- PMCID: PMC8648559
- DOI: 10.1038/s41592-021-01326-w
OME-NGFF: a next-generation file format for expanding bioimaging data-access strategies
Abstract
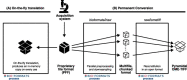
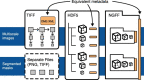
The rapid pace of innovation in biological imaging and the diversity of its applications have prevented the establishment of a community-agreed standardized data format. We propose that complementing established open formats such as OME-TIFF and HDF5 with a next-generation file format such as Zarr will satisfy the majority of use cases in bioimaging. Critically, a common metadata format used in all these vessels can deliver truly findable, accessible, interoperable and reusable bioimaging data.
© 2021. The Author(s).
Conflict of interest statement
C.A., E.D., K.K., M.L. and J.R.S. are affiliated with Glencoe Software, a commercial company that builds, delivers, supports and integrates image data management systems across academic, biotech and pharmaceutical industries. The remaining authors declare no competing interests.
Figures



Similar articles
-
OME-Zarr: a cloud-optimized bioimaging file format with international community support.Histochem Cell Biol. 2023 Sep;160(3):223-251. doi: 10.1007/s00418-023-02209-1. Epub 2023 Jul 10. Histochem Cell Biol. 2023. PMID: 37428210 Free PMC article.
-
Toward scalable reuse of vEM data: OME-Zarr to the rescue.Methods Cell Biol. 2023;177:359-387. doi: 10.1016/bs.mcb.2023.01.016. Epub 2023 Mar 9. Methods Cell Biol. 2023. PMID: 37451774
-
Metadata matters: access to image data in the real world.J Cell Biol. 2010 May 31;189(5):777-82. doi: 10.1083/jcb.201004104. J Cell Biol. 2010. PMID: 20513764 Free PMC article.
-
Common file formats.Curr Protoc Bioinformatics. 2007 Jan;Appendix 1:Appendix 1B. doi: 10.1002/0471250953.bia01bs16. Curr Protoc Bioinformatics. 2007. PMID: 18428774 Review.
-
Interoperability with Moby 1.0--it's better than sharing your toothbrush!Brief Bioinform. 2008 May;9(3):220-31. doi: 10.1093/bib/bbn003. Epub 2008 Jan 31. Brief Bioinform. 2008. PMID: 18238804 Review.
Cited by
-
SciJava Ops: an improved algorithms framework for Fiji and beyond.Front Bioinform. 2024 Sep 27;4:1435733. doi: 10.3389/fbinf.2024.1435733. eCollection 2024. Front Bioinform. 2024. PMID: 39399098 Free PMC article.
-
Arkitekt: streaming analysis and real-time workflows for microscopy.Nat Methods. 2024 Oct;21(10):1884-1894. doi: 10.1038/s41592-024-02404-5. Epub 2024 Sep 18. Nat Methods. 2024. PMID: 39294366
-
Ultrack: pushing the limits of cell tracking across biological scales.bioRxiv [Preprint]. 2024 Sep 3:2024.09.02.610652. doi: 10.1101/2024.09.02.610652. bioRxiv. 2024. PMID: 39282368 Free PMC article. Preprint.
-
Navigate: an open-source platform for smart light-sheet microscopy.Nat Methods. 2024 Sep 11. doi: 10.1038/s41592-024-02413-4. Online ahead of print. Nat Methods. 2024. PMID: 39261640 No abstract available.
-
BIOMERO: A scalable and extensible image analysis framework.Patterns (N Y). 2024 Jul 18;5(8):101024. doi: 10.1016/j.patter.2024.101024. eCollection 2024 Aug 9. Patterns (N Y). 2024. PMID: 39233696 Free PMC article.
References
-
- The HDF5 Library and File Format (The HDF Group, accessed 18 October 2021); https://www.hdfgroup.org/solutions/hdf5/
-
- Miles, A. et al. zarr-developers/zarr-python: v.2.5.0 10.5281/zenodo.4069231 (2020).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

