Population analysis of retrotransposons in giraffe genomes supports RTE decline and widespread LINE1 activity in Giraffidae
- PMID: 34836553
- PMCID: PMC8620236
- DOI: 10.1186/s13100-021-00254-y
Population analysis of retrotransposons in giraffe genomes supports RTE decline and widespread LINE1 activity in Giraffidae
Abstract
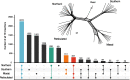
Background: The majority of structural variation in genomes is caused by insertions of transposable elements (TEs). In mammalian genomes, the main TE fraction is made up of autonomous and non-autonomous non-LTR retrotransposons commonly known as LINEs and SINEs (Long and Short Interspersed Nuclear Elements). Here we present one of the first population-level analysis of TE insertions in a non-model organism, the giraffe. Giraffes are ruminant artiodactyls, one of the few mammalian groups with genomes that are colonized by putatively active LINEs of two different clades of non-LTR retrotransposons, namely the LINE1 and RTE/BovB LINEs as well as their associated SINEs. We analyzed TE insertions of both types, and their associated SINEs in three giraffe genome assemblies, as well as across a population level sampling of 48 individuals covering all extant giraffe species.
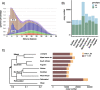
Results: The comparative genome screen identified 139,525 recent LINE1 and RTE insertions in the sampled giraffe population. The analysis revealed a drastically reduced RTE activity in giraffes, whereas LINE1 is still actively propagating in the genomes of extant (sub)-species. In concert with the extremely low activity of the giraffe RTE, we also found that RTE-dependent SINEs, namely Bov-tA and Bov-A2, have been virtually immobile in the last 2 million years. Despite the high current activity of the giraffe LINE1, we did not find evidence for the presence of currently active LINE1-dependent SINEs. TE insertion heterozygosity rates differ among the different (sub)-species, likely due to divergent population histories.
Conclusions: The horizontally transferred RTE/BovB and its derived SINEs appear to be close to inactivation and subsequent extinction in the genomes of extant giraffe species. This is the first time that the decline of a TE family has been meticulously analyzed from a population genetics perspective. Our study shows how detailed information about past and present TE activity can be obtained by analyzing large-scale population-level genomic data sets.
Keywords: BovB; Genome; Giraffe; LINE; LINE1/L1; Non-LTR retrotransposons; RTE; Ruminantia; SINE; Structural variation; TE; Transposable elements.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
A genome survey sequencing of the Java mouse deer (Tragulus javanicus) adds new aspects to the evolution of lineage specific retrotransposons in Ruminantia (Cetartiodactyla).Gene. 2015 Oct 25;571(2):271-8. doi: 10.1016/j.gene.2015.06.064. Epub 2015 Jun 27. Gene. 2015. PMID: 26123917
-
Comparative analysis of transposable elements provides insights into genome evolution in the genus Camelus.BMC Genomics. 2021 Nov 20;22(1):842. doi: 10.1186/s12864-021-08117-9. BMC Genomics. 2021. PMID: 34800971 Free PMC article.
-
Hagfish genome reveals parallel evolution of 7SL RNA-derived SINEs.Mob DNA. 2020 May 22;11:18. doi: 10.1186/s13100-020-00210-2. eCollection 2020. Mob DNA. 2020. PMID: 32489435 Free PMC article.
-
[Non-LTR retrotransposons: LINEs and SINEs in plant genome].Yi Chuan. 2006 Jun;28(6):731-6. Yi Chuan. 2006. PMID: 16818439 Review. Chinese.
-
LINEs, SINEs and repetitive DNA: non-LTR retrotransposons in plant genomes.Plant Mol Biol. 1999 Aug;40(6):903-10. doi: 10.1023/a:1006212929794. Plant Mol Biol. 1999. PMID: 10527415 Review.
Cited by
-
A sheep pangenome reveals the spectrum of structural variations and their effects on tail phenotypes.Genome Res. 2023 Mar;33(3):463-477. doi: 10.1101/gr.277372.122. Epub 2023 Mar 27. Genome Res. 2023. PMID: 37310928 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

