HIV-1 Envelope Glycoproteins Proteolytic Cleavage Protects Infected Cells from ADCC Mediated by Plasma from Infected Individuals
- PMID: 34835042
- PMCID: PMC8625184
- DOI: 10.3390/v13112236
HIV-1 Envelope Glycoproteins Proteolytic Cleavage Protects Infected Cells from ADCC Mediated by Plasma from Infected Individuals
Abstract
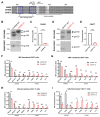
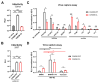
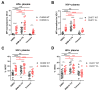
The HIV-1 envelope glycoprotein (Env) is synthesized in the endoplasmic reticulum as a trimeric gp160 precursor, which requires proteolytic cleavage by a cellular furin protease to mediate virus-cell fusion. Env is conformationally flexible but controls its transition from the unbound "closed" conformation (State 1) to downstream CD4-bound conformations (States 2/3), which are required for fusion. In particular, HIV-1 has evolved several mechanisms that reduce the premature "opening" of Env which exposes highly conserved epitopes recognized by non-neutralizing antibodies (nnAbs) capable of mediating antibody-dependent cellular cytotoxicity (ADCC). Env cleavage decreases its conformational transitions favoring the adoption of the "closed" conformation. Here we altered the gp160 furin cleavage site to impair Env cleavage and to examine its impact on ADCC responses mediated by plasma from HIV-1-infected individuals. We found that infected primary CD4+ T cells expressing uncleaved, but not wildtype, Env are efficiently recognized by nnAbs and become highly susceptible to ADCC responses mediated by plasma from HIV-1-infected individuals. Thus, HIV-1 limits the exposure of uncleaved Env at the surface of HIV-1-infected cells at least in part to escape ADCC responses.
Keywords: ADCC; CD4 mimetics; Env glycoprotein; HIV+ plasma; HIV-1; Temsavir; furin cleavage site; nnAbs.
Conflict of interest statement
The authors declare no competing interest.
Figures
Similar articles
-
CD4 downregulation precedes Env expression and protects HIV-1-infected cells from ADCC mediated by non-neutralizing antibodies.mBio. 2024 Nov 13;15(11):e0182724. doi: 10.1128/mbio.01827-24. Epub 2024 Oct 7. mBio. 2024. PMID: 39373535 Free PMC article.
-
Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity.J Virol. 2014 Mar;88(5):2633-44. doi: 10.1128/JVI.03230-13. Epub 2013 Dec 18. J Virol. 2014. PMID: 24352444 Free PMC article.
-
Co-receptor Binding Site Antibodies Enable CD4-Mimetics to Expose Conserved Anti-cluster A ADCC Epitopes on HIV-1 Envelope Glycoproteins.EBioMedicine. 2016 Oct;12:208-218. doi: 10.1016/j.ebiom.2016.09.004. Epub 2016 Sep 9. EBioMedicine. 2016. PMID: 27633463 Free PMC article.
-
Impact of HIV-1 Envelope Conformation on ADCC Responses.Trends Microbiol. 2018 Apr;26(4):253-265. doi: 10.1016/j.tim.2017.10.007. Epub 2017 Nov 20. Trends Microbiol. 2018. PMID: 29162391 Review.
-
Unlocking HIV-1 Env: implications for antibody attack.AIDS Res Ther. 2017 Sep 12;14(1):42. doi: 10.1186/s12981-017-0168-5. AIDS Res Ther. 2017. PMID: 28893275 Free PMC article. Review.
Cited by
-
HIV-1 Vpu restricts Fc-mediated effector functions in vivo.Cell Rep. 2022 Nov 8;41(6):111624. doi: 10.1016/j.celrep.2022.111624. Cell Rep. 2022. PMID: 36351384 Free PMC article.
-
Temsavir Treatment of HIV-1-Infected Cells Decreases Envelope Glycoprotein Recognition by Broadly Neutralizing Antibodies.mBio. 2022 Jun 28;13(3):e0057722. doi: 10.1128/mbio.00577-22. Epub 2022 Apr 27. mBio. 2022. PMID: 35475646 Free PMC article.
-
Temsavir Modulates HIV-1 Envelope Conformation by Decreasing Its Proteolytic Cleavage.Viruses. 2023 May 18;15(5):1189. doi: 10.3390/v15051189. Viruses. 2023. PMID: 37243275 Free PMC article.
References
-
- Qi M., Williams J.A., Chu H., Chen X., Wang J.J., Ding L., Akhirome E., Wen X., Lapierre L.A., Goldenring J.R., et al. Rab11-FIP1C and Rab14 direct plasma membrane sorting and particle incorporation of the HIV-1 envelope glycoprotein complex. PLoS Pathog. 2013;9:e1003278. doi: 10.1371/journal.ppat.1003278. - DOI - PMC - PubMed
-
- Kirschman J., Qi M., Ding L., Hammonds J., Dienger-Stambaugh K., Wang J.J., Lapierre L.A., Goldenring J.R., Spearman P. HIV-1 Envelope Glycoprotein Trafficking through the Endosomal Recycling Compartment Is Required for Particle Incorporation. J. Virol. 2018;92 doi: 10.1128/JVI.01893-17. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

