Insertion of Exogenous Genes within the ORF1a Coding Region of Porcine Astrovirus
- PMID: 34834925
- PMCID: PMC8623754
- DOI: 10.3390/v13112119
Insertion of Exogenous Genes within the ORF1a Coding Region of Porcine Astrovirus
Abstract
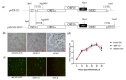
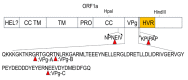
A tagged or reporter astrovirus can be a valuable tool for the analysis of various aspects of the virus life cycle, and to aid in the development of genetically engineered astroviruses as vectors. Here, transposon-mediated insertion mutagenesis was used to insert a 15-nucleotide (nt) sequence into random sites of open reading frame 1a (ORF1a) based on an infectious full-length cDNA clone of porcine astrovirus (PAstV). Five sites in the predicted coiled-coil structures (CC), genome-linked protein (VPg), and hypervariable region (HVR) in ORF1a of the PAstV genome were identified that could tolerate random 15 nt insertions. Incorporation of the commonly used epitope tags, His, Flag, and HA, into four of the five insertion sites permitted the production of infectious viruses and allowed recognition by specifically tagged monoclonal antibodies. The results of immuno-fluorescent assays showed that Flag-tagged ORF1a protein overlapped partially with capsid and ORF2b proteins in the cytoplasm. Improved light-oxygen-voltage (iLOV) gene was also introduced at the insertion sites of CC, VPg, and HVR. Only one viable recombinant reporter PAstV expressing iLOV inserted in HVR was recovered. Biological analysis of the reporter virus showed that it displayed similar growth characteristics, and yet produced less infectious virus particles, when compared with the parental virus. The recombinant virus carrying the iLOV fused with the HVR of ORF1a protein maintained its stability and showed green fluorescence after 15 passages in cell cultures. The resultant fluorescently tagged virus could provide a promising tool for the rapid screening of antiviral drugs as well as allowing the visualization of PAstV infection and replication in living cells.
Keywords: DNA-launched infectious clones; HVR; PASTV-GX1; astrovirus; iLOV; tag; transposons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Insertion of exogenous genes within the ORF1b coding region of porcine astrovirus.Vet Microbiol. 2023 May;280:109675. doi: 10.1016/j.vetmic.2023.109675. Epub 2023 Feb 10. Vet Microbiol. 2023. PMID: 36812864
-
Complete genome sequence of a porcine astrovirus from South Korea.Arch Virol. 2015 Jul;160(7):1819-21. doi: 10.1007/s00705-015-2436-9. Epub 2015 Apr 29. Arch Virol. 2015. PMID: 25916612
-
Comparative Analysis of Novel Strains of Porcine Astrovirus Type 3 in the USA.Viruses. 2021 Sep 17;13(9):1859. doi: 10.3390/v13091859. Viruses. 2021. PMID: 34578440 Free PMC article.
-
The molecular biology of astroviruses.Arch Virol Suppl. 1996;12:277-85. doi: 10.1007/978-3-7091-6553-9_30. Arch Virol Suppl. 1996. PMID: 9015125 Review.
-
Molecular biology of astroviruses: selected highlights.Novartis Found Symp. 2001;238:219-33; discussion 233-6. doi: 10.1002/0470846534.ch13. Novartis Found Symp. 2001. PMID: 11444028 Review.
References
-
- Fernández-Correa I., Truchado D.A., Gomez A.D., Doménech A., Pérez-Tris J., Schmidt-Chanasit J., Cadar D., Benítez L. A novel group of avian astroviruses from Neotropical passerine birds broaden the diversity and host range of Astroviridae. Sci. Rep. 2019;9:9513. doi: 10.1038/s41598-019-45889-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials

