Reactivities of a Prostanoid EP2 Agonist, Omidenepag, Are Useful for Distinguishing between 3D Spheroids of Human Orbital Fibroblasts without or with Graves' Orbitopathy
- PMID: 34831419
- PMCID: PMC8622545
- DOI: 10.3390/cells10113196
Reactivities of a Prostanoid EP2 Agonist, Omidenepag, Are Useful for Distinguishing between 3D Spheroids of Human Orbital Fibroblasts without or with Graves' Orbitopathy
Abstract
Background: To obtain new insights into the activation of the thyroid-stimulating hormone (TSH) and insulin-like growth factor 1 (IGF-1) receptors in human orbital fibroblasts (n-HOFs), the effects of the prostanoid EP2 agonist, omidenepag (OMD), and a rho-associated coiled-coil-containing protein kinase (ROCK) inhibitor, ripasudil (Rip) were evaluated using three-dimension (3D) n-HOFs spheroids in the absence and presence of the recombinant human TSH receptor antibodies, M22 and IGF-1.
Methods: The effects of 100 nM OMD or 10 μM Rip on the physical properties, size, stiffness, and mRNA expression of several extracellular matrix (ECM) molecules, their regulator, inflammatory cytokines, and endoplasmic reticulum (ER) stress-related factors were examined and compared among 3D spheroids of n-HOFs, M22-/IGF-1-activated n-HOFs and GO-related human orbital fibroblasts (GHOFs).
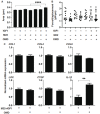
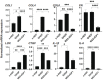
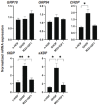
Results: The physical properties and mRNA expressions of several genes of the 3D n-HOFs spheroids were significantly and diversely modulated by the presence of OMD or Rip. The OMD-induced effects on M22-/IGF-1-activated n-HOFs were similar to the effects caused by GHOHs, but quite different from those of n-HOFs.
Conclusions: The findings presented herein indicate that the changes induced by OMD may be useful in distinguishing between n-HOFs and GHOFs.
Keywords: Graves’ orbitopathy; IGF-1; ROCK inhibitor; orbital fibroblast; prostanoid EP2 agonist; three-dimension (3D) cell culture.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Prostaglandin F2α and EP2 agonists, and a ROCK inhibitor modulate the formation of 3D organoids of Grave's orbitopathy related human orbital fibroblasts.Exp Eye Res. 2021 Apr;205:108489. doi: 10.1016/j.exer.2021.108489. Epub 2021 Feb 12. Exp Eye Res. 2021. PMID: 33587909
-
Effects of linsitinib on M22 and IGF:1-treated 3D spheroids of human orbital fibroblasts.Sci Rep. 2025 Jan 2;15(1):384. doi: 10.1038/s41598-024-83193-x. Sci Rep. 2025. PMID: 39747159 Free PMC article.
-
Simultaneous Use of ROCK Inhibitors and EP2 Agonists Induces Unexpected Effects on Adipogenesis and the Physical Properties of 3T3-L1 Preadipocytes.Int J Mol Sci. 2021 Apr 28;22(9):4648. doi: 10.3390/ijms22094648. Int J Mol Sci. 2021. PMID: 33925005 Free PMC article.
-
Current perspectives on the role of orbital fibroblasts in the pathogenesis of Graves' ophthalmopathy.Exp Eye Res. 2016 Jan;142:83-91. doi: 10.1016/j.exer.2015.02.007. Exp Eye Res. 2016. PMID: 26675405 Review.
-
Role of insulin-like growth factor-1 (IGF-1) pathway in the pathogenesis of Graves' orbitopathy.Best Pract Res Clin Endocrinol Metab. 2012 Jun;26(3):291-302. doi: 10.1016/j.beem.2011.10.002. Best Pract Res Clin Endocrinol Metab. 2012. PMID: 22632366 Free PMC article. Review.
Cited by
-
Application of Single Cell Type-Derived Spheroids Generated by Using a Hanging Drop Culture Technique in Various In Vitro Disease Models: A Narrow Review.Cells. 2024 Sep 14;13(18):1549. doi: 10.3390/cells13181549. Cells. 2024. PMID: 39329734 Free PMC article. Review.
-
Physical Properties and Cellular Metabolic Characteristics of 3D Spheroids Are Possible Definitive Indices for the Biological Nature of Cancer-Associated Fibroblasts.Cells. 2023 Aug 28;12(17):2160. doi: 10.3390/cells12172160. Cells. 2023. PMID: 37681892 Free PMC article.
-
Unexpected Crosslinking Effects of a Human Thyroid Stimulating Monoclonal Autoantibody, M22, with IGF1 on Adipogenesis in 3T3L-1 Cells.Int J Mol Sci. 2023 Jan 6;24(2):1110. doi: 10.3390/ijms24021110. Int J Mol Sci. 2023. PMID: 36674625 Free PMC article.
References
-
- Turcu A.F., Kumar S., Neumann S., Coenen M., Iyer S., Chiriboga P., Gershengorn M.C., Bahn R.S. A small molecule antagonist inhibits thyrotropin receptor antibody-induced orbital fibroblast functions involved in the pathogenesis of Graves ophthalmopathy. J. Clin. Endocrinol. Metab. 2013;98:2153–2159. doi: 10.1210/jc.2013-1149. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

