Evolution of the Early Spliceosomal Complex-From Constitutive to Regulated Splicing
- PMID: 34830325
- PMCID: PMC8624252
- DOI: 10.3390/ijms222212444
Evolution of the Early Spliceosomal Complex-From Constitutive to Regulated Splicing
Abstract
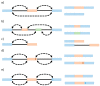
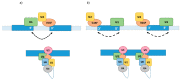
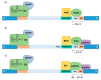
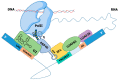
Pre-mRNA splicing is a major process in the regulated expression of genes in eukaryotes, and alternative splicing is used to generate different proteins from the same coding gene. Splicing is a catalytic process that removes introns and ligates exons to create the RNA sequence that codifies the final protein. While this is achieved in an autocatalytic process in ancestral group II introns in prokaryotes, the spliceosome has evolved during eukaryogenesis to assist in this process and to finally provide the opportunity for intron-specific splicing. In the early stage of splicing, the RNA 5' and 3' splice sites must be brought within proximity to correctly assemble the active spliceosome and perform the excision and ligation reactions. The assembly of this first complex, termed E-complex, is currently the least understood process. We focused in this review on the formation of the E-complex and compared its composition and function in three different organisms. We highlight the common ancestral mechanisms in S. cerevisiae, S. pombe, and mammals and conclude with a unifying model for intron definition in constitutive and regulated co-transcriptional splicing.
Keywords: 5′ splicing site; E-complex; Prp2; U2AF65; exon–intron junction; fission yeast; spliceosome; splicing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The NineTeen Complex (NTC) and NTC-associated proteins as targets for spliceosomal ATPase action during pre-mRNA splicing.RNA Biol. 2015;12(2):109-14. doi: 10.1080/15476286.2015.1008926. RNA Biol. 2015. PMID: 25654271 Free PMC article.
-
The evolutionary conservation of the splicing apparatus between fission yeast and man.Nucleic Acids Symp Ser. 1995;(33):226-8. Nucleic Acids Symp Ser. 1995. PMID: 8643378 Review.
-
Interactions of SR45, an SR-like protein, with spliceosomal proteins and an intronic sequence: insights into regulated splicing.Plant J. 2012 Sep;71(6):936-47. doi: 10.1111/j.1365-313X.2012.05042.x. Epub 2012 Jun 28. Plant J. 2012. PMID: 22563826
-
Genome-wide analysis of pre-mRNA splicing: intron features govern the requirement for the second-step factor, Prp17 in Saccharomyces cerevisiae and Schizosaccharomyces pombe.J Biol Chem. 2004 Dec 10;279(50):52437-46. doi: 10.1074/jbc.M408815200. Epub 2004 Sep 27. J Biol Chem. 2004. PMID: 15452114
-
How Is Precursor Messenger RNA Spliced by the Spliceosome?Annu Rev Biochem. 2020 Jun 20;89:333-358. doi: 10.1146/annurev-biochem-013118-111024. Epub 2019 Dec 9. Annu Rev Biochem. 2020. PMID: 31815536 Review.
Cited by
-
Intramolecular autoinhibition regulates the selectivity of PRPF40A tandem WW domains for proline-rich motifs.Nat Commun. 2024 May 8;15(1):3888. doi: 10.1038/s41467-024-48004-x. Nat Commun. 2024. PMID: 38719828 Free PMC article.
-
Commitment Complex Splicing Factors in Cancers of the Gastrointestinal Tract-An In Silico Study.Bioinform Biol Insights. 2024 Oct 15;18:11779322241287115. doi: 10.1177/11779322241287115. eCollection 2024. Bioinform Biol Insights. 2024. PMID: 39421280 Free PMC article.
-
Evidence-Based Guide to Using Artificial Introns for Tissue-Specific Knockout in Mice.Int J Mol Sci. 2023 Jun 17;24(12):10258. doi: 10.3390/ijms241210258. Int J Mol Sci. 2023. PMID: 37373404 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

