Beyond HAT Adaptor: TRRAP Liaisons with Sp1-Mediated Transcription
- PMID: 34830324
- PMCID: PMC8625110
- DOI: 10.3390/ijms222212445
Beyond HAT Adaptor: TRRAP Liaisons with Sp1-Mediated Transcription
Abstract
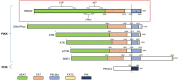
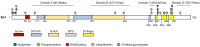
The members of the phosphatidylinositol 3-kinase-related kinase (PIKK) family play vital roles in multiple biological processes, including DNA damage response, metabolism, cell growth, mRNA decay, and transcription. TRRAP, as the only member lacking the enzymatic activity in this family, is an adaptor protein for several histone acetyltransferase (HAT) complexes and a scaffold protein for multiple transcription factors. TRRAP has been demonstrated to regulate various cellular functions in cell cycle progression, cell stemness maintenance and differentiation, as well as neural homeostasis. TRRAP is known to be an important orchestrator of many molecular machineries in gene transcription by modulating the activity of some key transcription factors, including E2F1, c-Myc, p53, and recently, Sp1. This review summarizes the biological and biochemical studies on the action mode of TRRAP together with the transcription factors, focusing on how TRRAP-HAT mediates the transactivation of Sp1-governing biological processes, including neurodegeneration.
Keywords: HAT; Sp1; TRRAP; neuodegeneration; neuro-development; transcription.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
HAT cofactor TRRAP modulates microtubule dynamics via SP1 signaling to prevent neurodegeneration.Elife. 2021 Feb 17;10:e61531. doi: 10.7554/eLife.61531. Elife. 2021. PMID: 33594975 Free PMC article.
-
MYC interacts with the human STAGA coactivator complex via multivalent contacts with the GCN5 and TRRAP subunits.Biochim Biophys Acta. 2014 May;1839(5):395-405. doi: 10.1016/j.bbagrm.2014.03.017. Epub 2014 Apr 3. Biochim Biophys Acta. 2014. PMID: 24705139 Free PMC article.
-
Orchestration of chromatin-based processes: mind the TRRAP.Oncogene. 2007 Aug 13;26(37):5358-72. doi: 10.1038/sj.onc.1210605. Oncogene. 2007. PMID: 17694078 Review.
-
An hGCN5/TRRAP histone acetyltransferase complex co-activates BRCA1 transactivation function through histone modification.J Biol Chem. 2006 Jan 6;281(1):20-6. doi: 10.1074/jbc.M510157200. Epub 2005 Oct 31. J Biol Chem. 2006. Retraction in: J Biol Chem. 2013 Oct 18;288(42):30507. doi: 10.1074/jbc.A113.510157 PMID: 16260778 Retracted.
-
New insights into the evolutionary conservation of the sole PIKK pseudokinase Tra1/TRRAP.Biochem Soc Trans. 2019 Dec 20;47(6):1597-1608. doi: 10.1042/BST20180496. Biochem Soc Trans. 2019. PMID: 31769470 Review.
Cited by
-
SP1 undergoes phase separation and activates RGS20 expression through super-enhancers to promote lung adenocarcinoma progression.Proc Natl Acad Sci U S A. 2024 Jul 16;121(29):e2401834121. doi: 10.1073/pnas.2401834121. Epub 2024 Jul 8. Proc Natl Acad Sci U S A. 2024. PMID: 38976739 Free PMC article.
-
Histone and Histone Acetylation-Related Alterations of Gene Expression in Uninvolved Psoriatic Skin and Their Effects on Cell Proliferation, Differentiation, and Immune Responses.Int J Mol Sci. 2023 Sep 26;24(19):14551. doi: 10.3390/ijms241914551. Int J Mol Sci. 2023. PMID: 37833997 Free PMC article.
-
Exploring the Genetic Predisposition to Epigenetic Changes in Alzheimer's Disease.Int J Mol Sci. 2023 Apr 27;24(9):7955. doi: 10.3390/ijms24097955. Int J Mol Sci. 2023. PMID: 37175659 Free PMC article.
-
Unraveling the Behavior of Intrinsically Disordered Protein c-Myc: A Study Utilizing Gaussian-Accelerated Molecular Dynamics.ACS Omega. 2023 Dec 1;9(2):2250-2262. doi: 10.1021/acsomega.3c05822. eCollection 2024 Jan 16. ACS Omega. 2023. PMID: 38250404 Free PMC article.
-
Long non-coding RNAs and microRNAs as crucial regulators in cardio-oncology.Cell Biosci. 2022 Mar 4;12(1):24. doi: 10.1186/s13578-022-00757-y. Cell Biosci. 2022. PMID: 35246252 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

