Canonical and Divergent N-Terminal HBx Isoform Proteins Unveiled: Characteristics and Roles during HBV Replication
- PMID: 34829930
- PMCID: PMC8616016
- DOI: 10.3390/biomedicines9111701
Canonical and Divergent N-Terminal HBx Isoform Proteins Unveiled: Characteristics and Roles during HBV Replication
Abstract
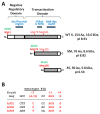
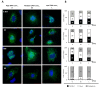
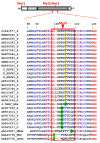
Hepatitis B virus (HBV) X protein (HBx) is a viral regulatory and multifunctional protein. It is well-known that the canonical HBx reading frame bears two phylogenetically conserved internal in-frame translational initiation codons at Met2 and Met3, thus possibly generating divergent N-terminal smaller isoforms during translation. Here, we demonstrate that the three distinct HBx isoforms are generated from the ectopically expressed HBV HBx gene, named XF (full-length), XM (medium-length), and XS (short-length); they display different subcellular localizations when expressed individually in cultured hepatoma cells. Particularly, the smallest HBx isoform, XS, displayed a predominantly cytoplasmic localization. To study HBx proteins during viral replication, we performed site-directed mutagenesis to target the individual or combinatorial expression of the HBx isoforms within the HBV viral backbone (full viral genome). Our results indicate that of all HBx isoforms, only the smallest HBx isoform, XS, can restore WT levels of HBV replication, and bind to the viral mini chromosome, thereby establishing an active chromatin state, highlighting its crucial activities during HBV replication. Intriguingly, we found that sequences of HBV HBx genotype H are devoid of the conserved Met3 position, and therefore HBV genotype H infection is naturally silent for the expression of the HBx XS isoform. Finally, we found that the HBx XM (medium-length) isoform shares significant sequence similarity with the N-terminus domain of the COMMD8 protein, a member of the copper metabolism MURR1 domain-containing (COMMD) protein family. This novel finding might facilitate studies on the phylogenetic origin of the HBV X protein. The identification and functional characterization of its isoforms will shift the paradigm by changing the concept of HBx from being a unique, canonical, and multifunctional protein toward the occurrence of different HBx isoforms, carrying out different overlapping functions at different subcellular localizations during HBV genome replication. Significantly, our current work unveils new crucial HBV targets to study for potential antiviral research, and human virus pathogenesis.
Keywords: HBV; HBx; divergent N-terminal isoform; genome replication; hepatitis B virus; hepatitis B virus X protein; localization regulation; subcellular localization.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Pre- and Post-Transcriptional Control of HBV Gene Expression: The Road Traveled towards the New Paradigm of HBx, Its Isoforms, and Their Diverse Functions.Biomedicines. 2023 Jun 9;11(6):1674. doi: 10.3390/biomedicines11061674. Biomedicines. 2023. PMID: 37371770 Free PMC article. Review.
-
Phosphorylation of Phylogenetically Conserved Amino Acid Residues Confines HBx within Different Cell Compartments of Human Hepatocarcinoma Cells.Molecules. 2021 Feb 26;26(5):1254. doi: 10.3390/molecules26051254. Molecules. 2021. PMID: 33652602 Free PMC article.
-
Smc5/6 Antagonism by HBx Is an Evolutionarily Conserved Function of Hepatitis B Virus Infection in Mammals.J Virol. 2018 Jul 31;92(16):e00769-18. doi: 10.1128/JVI.00769-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29848586 Free PMC article.
-
Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death.J Virol. 2005 Apr;79(7):4238-45. doi: 10.1128/JVI.79.7.4238-4245.2005. J Virol. 2005. PMID: 15767425 Free PMC article.
-
Hepatitis B Virus X and Regulation of Viral Gene Expression.Cold Spring Harb Perspect Med. 2016 Jan 8;6(3):a021402. doi: 10.1101/cshperspect.a021402. Cold Spring Harb Perspect Med. 2016. PMID: 26747833 Free PMC article. Review.
Cited by
-
Pre- and Post-Transcriptional Control of HBV Gene Expression: The Road Traveled towards the New Paradigm of HBx, Its Isoforms, and Their Diverse Functions.Biomedicines. 2023 Jun 9;11(6):1674. doi: 10.3390/biomedicines11061674. Biomedicines. 2023. PMID: 37371770 Free PMC article. Review.
-
Genetic Diversity of Hepatitis B and C Viruses Revealed by Continuous Surveillance from 2015 to 2021 in Gabon, Central Africa.Microorganisms. 2023 Aug 9;11(8):2046. doi: 10.3390/microorganisms11082046. Microorganisms. 2023. PMID: 37630606 Free PMC article.
-
Relevance of HBx for Hepatitis B Virus-Associated Pathogenesis.Int J Mol Sci. 2023 Mar 4;24(5):4964. doi: 10.3390/ijms24054964. Int J Mol Sci. 2023. PMID: 36902395 Free PMC article. Review.
References
-
- WHO Global Hepatitis Report, 2017. 2017. [(accessed on 15 June 2021)]. Available online: https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/
LinkOut - more resources
Full Text Sources

