RadA, a MSCRAMM Adhesin of the Dominant Symbiote Ruminococcus gnavus E1, Binds Human Immunoglobulins and Intestinal Mucins
- PMID: 34827611
- PMCID: PMC8615915
- DOI: 10.3390/biom11111613
RadA, a MSCRAMM Adhesin of the Dominant Symbiote Ruminococcus gnavus E1, Binds Human Immunoglobulins and Intestinal Mucins
Abstract
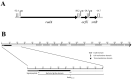
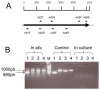
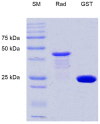
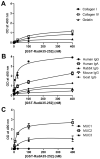
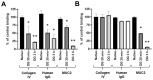
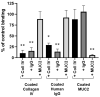
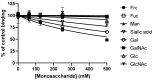
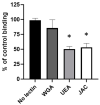
Adhesion to the digestive mucosa is considered a key factor for bacterial persistence within the gut. In this study, we show that Ruminococcus gnavus E1 can express the radA gene, which encodes an adhesin of the MSCRAMMs family, only when it colonizes the gut. The RadA N-terminal region contains an all-β bacterial Ig-like domain known to interact with collagens. We observed that it preferentially binds human immunoglobulins (IgA and IgG) and intestinal mucins. Using deglycosylated substrates, we also showed that the RadA N-terminal region recognizes two different types of motifs, the protein backbone of human IgG and the glycan structure of mucins. Finally, competition assays with lectins and free monosaccharides identified Galactose and N-Acetyl-Galactosamine motifs as specific targets for the binding of RadA to mucins and the surface of human epithelial cells.
Keywords: Caco-2; HT-29-16E; Ruminococcus gnavus; adhesin; bacterial Ig-like domain; collagen; mucin; mucus; solid phase assay.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Unravelling the specificity and mechanism of sialic acid recognition by the gut symbiont Ruminococcus gnavus.Nat Commun. 2017 Dec 19;8(1):2196. doi: 10.1038/s41467-017-02109-8. Nat Commun. 2017. PMID: 29259165 Free PMC article.
-
Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent.PLoS One. 2013 Oct 25;8(10):e76341. doi: 10.1371/journal.pone.0076341. eCollection 2013. PLoS One. 2013. PMID: 24204617 Free PMC article.
-
The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging: Implication for niche colonisation in the gastrointestinal tract.PLoS Biol. 2021 Dec 22;19(12):e3001498. doi: 10.1371/journal.pbio.3001498. eCollection 2021 Dec. PLoS Biol. 2021. PMID: 34936658 Free PMC article.
-
How do they stick together? Bacterial adhesins implicated in the binding of bacteria to the human gastrointestinal mucins.Biochem Soc Trans. 2017 Apr 15;45(2):389-399. doi: 10.1042/BST20160167. Biochem Soc Trans. 2017. PMID: 28408479 Review.
-
Structural insights into bacterial recognition of intestinal mucins.Curr Opin Struct Biol. 2014 Oct;28:23-31. doi: 10.1016/j.sbi.2014.07.002. Epub 2014 Aug 6. Curr Opin Struct Biol. 2014. PMID: 25106027 Review.
Cited by
-
A mucin-regulated adhesin determines the spatial organization and inflammatory character of a bacterial symbiont in the vertebrate gut.Cell Host Microbe. 2023 Aug 9;31(8):1371-1385.e6. doi: 10.1016/j.chom.2023.07.003. Epub 2023 Jul 28. Cell Host Microbe. 2023. PMID: 37516109 Free PMC article.
-
Ruminococcus gnavus: friend or foe for human health.FEMS Microbiol Rev. 2023 Mar 10;47(2):fuad014. doi: 10.1093/femsre/fuad014. FEMS Microbiol Rev. 2023. PMID: 37015876 Free PMC article. Review.
-
Supplementation with honeysuckle extract improves growth performance, immune performance, gut morphology, and cecal microbes in geese.Front Vet Sci. 2022 Nov 3;9:1006318. doi: 10.3389/fvets.2022.1006318. eCollection 2022. Front Vet Sci. 2022. PMID: 36406074 Free PMC article.
-
Bacterial biofilms in the human body: prevalence and impacts on health and disease.Front Cell Infect Microbiol. 2023 Aug 30;13:1237164. doi: 10.3389/fcimb.2023.1237164. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37712058 Free PMC article. Review.
References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

