Interrogation of the microenvironmental landscape in spinal ependymomas reveals dual functions of tumor-associated macrophages
- PMID: 34824203
- PMCID: PMC8617028
- DOI: 10.1038/s41467-021-27018-9
Interrogation of the microenvironmental landscape in spinal ependymomas reveals dual functions of tumor-associated macrophages
Abstract
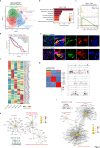
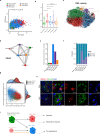
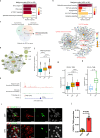
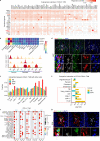
Spinal ependymomas are the most common spinal cord tumors in adults, but their intratumoral cellular heterogeneity has been less studied, and how spinal microglia are involved in tumor progression is still unknown. Here, our single-cell RNA-sequencing analyses of three spinal ependymoma subtypes dissect the microenvironmental landscape of spinal ependymomas and reveal tumor-associated macrophage (TAM) subsets with distinct functional phenotypes. CCL2+ TAMs are related to the immune response and exhibit a high capacity for apoptosis, while CD44+ TAMs are associated with tumor angiogenesis. By combining these results with those of single-cell ATAC-sequencing data analysis, we reveal that TEAD1 and EGR3 play roles in regulating the functional diversity of TAMs. We further identify diverse characteristics of both malignant cells and TAMs that might underlie the different malignant degrees of each subtype. Finally, assessment of cell-cell interactions reveal that stromal cells act as extracellular factors that mediate TAM diversity. Overall, our results reveal dual functions of TAMs in tumor progression, providing valuable insights for TAM-targeting immunotherapy.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Single-cell transcriptional atlas of tumor-associated macrophages in breast cancer.Breast Cancer Res. 2024 Sep 4;26(1):129. doi: 10.1186/s13058-024-01887-6. Breast Cancer Res. 2024. PMID: 39232806 Free PMC article.
-
Decoding the spatiotemporal heterogeneity of tumor-associated macrophages.Mol Cancer. 2024 Jul 27;23(1):150. doi: 10.1186/s12943-024-02064-1. Mol Cancer. 2024. PMID: 39068459 Free PMC article. Review.
-
Integrated analysis of single-cell RNA-seq and bulk RNA-seq unravels tumour heterogeneity plus M2-like tumour-associated macrophage infiltration and aggressiveness in TNBC.Cancer Immunol Immunother. 2021 Jan;70(1):189-202. doi: 10.1007/s00262-020-02669-7. Epub 2020 Jul 17. Cancer Immunol Immunother. 2021. PMID: 32681241 Free PMC article.
-
Single-cell transcriptomics reveal distinct immune-infiltrating phenotypes and macrophage-tumor interaction axes among different lineages of pituitary neuroendocrine tumors.Genome Med. 2024 Apr 24;16(1):60. doi: 10.1186/s13073-024-01325-4. Genome Med. 2024. PMID: 38658971 Free PMC article.
-
Advanced insights on tumor-associated macrophages revealed by single-cell RNA sequencing: The intratumor heterogeneity, functional phenotypes, and cellular interactions.Cancer Lett. 2024 Mar 1;584:216610. doi: 10.1016/j.canlet.2024.216610. Epub 2024 Jan 19. Cancer Lett. 2024. PMID: 38244910 Review.
Cited by
-
Reprogramming Tumor-Associated Macrophage Using Nanocarriers: New Perspectives to Halt Cancer Progression.Pharmaceutics. 2024 May 9;16(5):636. doi: 10.3390/pharmaceutics16050636. Pharmaceutics. 2024. PMID: 38794298 Free PMC article. Review.
-
Single cell analysis revealed SFRP2 cancer associated fibroblasts drive tumorigenesis in head and neck squamous cell carcinoma.NPJ Precis Oncol. 2024 Oct 9;8(1):228. doi: 10.1038/s41698-024-00716-5. NPJ Precis Oncol. 2024. PMID: 39384902 Free PMC article.
-
Metabolic regulation of tumor-associated macrophage heterogeneity: insights into the tumor microenvironment and immunotherapeutic opportunities.Biomark Res. 2024 Jan 7;12(1):1. doi: 10.1186/s40364-023-00549-7. Biomark Res. 2024. PMID: 38185636 Free PMC article. Review.
-
Tumor-associated macrophages: orchestrators of cholangiocarcinoma progression.Front Immunol. 2024 Sep 3;15:1451474. doi: 10.3389/fimmu.2024.1451474. eCollection 2024. Front Immunol. 2024. PMID: 39290697 Free PMC article. Review.
-
Heterogeneity of glioblastoma stem cells in the context of the immune microenvironment and geospatial organization.Front Oncol. 2022 Oct 19;12:1022716. doi: 10.3389/fonc.2022.1022716. eCollection 2022. Front Oncol. 2022. PMID: 36338705 Free PMC article. Review.
References
-
- Hanbali F, et al. Spinal cord ependymoma: radical surgical resection and outcome. Neurosurgery. 2002;51:1162–1172. - PubMed
-
- Connolly ID, Ali R, Li Y, Gephart MH. Genetic and molecular distinctions in spinal ependymomas: a review. Clin. Neurol. Neurosurg. 2015;139:210–215. - PubMed
-
- Ruda R, Gilbert M, Soffietti R. Ependymomas of the adult: molecular biology and treatment. Curr. Opin. Neurol. 2008;21:754–761. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

