Widespread microRNA degradation elements in target mRNAs can assist the encoded proteins
- PMID: 34819352
- PMCID: PMC8653786
- DOI: 10.1101/gad.348874.121
Widespread microRNA degradation elements in target mRNAs can assist the encoded proteins
Abstract
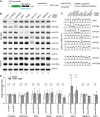
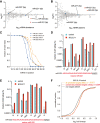
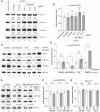
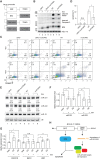
Binding of microRNAs (miRNAs) to mRNAs normally results in post-transcriptional repression of gene expression. However, extensive base-pairing between miRNAs and target RNAs can trigger miRNA degradation, a phenomenon called target RNA-directed miRNA degradation (TDMD). Here, we systematically analyzed Argonaute-CLASH (cross-linking, ligation, and sequencing of miRNA-target RNA hybrids) data and identified numerous candidate TDMD triggers, focusing on their ability to induce nontemplated nucleotide addition at the miRNA 3' end. When exogenously expressed in various cell lines, eight triggers induce degradation of corresponding miRNAs. Both the TDMD base-pairing and surrounding sequences are essential for TDMD. CRISPR knockout of endogenous trigger or ZSWIM8, a ubiquitin ligase essential for TDMD, reduced miRNA degradation. Furthermore, degradation of miR-221 and miR-222 by a trigger in BCL2L11, which encodes a proapoptotic protein, enhances apoptosis. Therefore, we uncovered widespread TDMD triggers in target RNAs and demonstrated an example that could functionally cooperate with the encoded protein.
Keywords: AGO-CLASH; BCL2L11; TDMD; apoptosis; microRNA.
© 2021 Li et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


Similar articles
-
Screening of Drosophila microRNA-degradation sequences reveals Argonaute1 mRNA's role in regulating miR-999.Nat Commun. 2023 Apr 13;14(1):2108. doi: 10.1038/s41467-023-37819-9. Nat Commun. 2023. PMID: 37055443 Free PMC article.
-
To kill a microRNA: emerging concepts in target-directed microRNA degradation.Nucleic Acids Res. 2024 Feb 28;52(4):1558-1574. doi: 10.1093/nar/gkae003. Nucleic Acids Res. 2024. PMID: 38224449 Free PMC article. Review.
-
The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation.Science. 2020 Dec 18;370(6523):eabc9359. doi: 10.1126/science.abc9359. Epub 2020 Nov 12. Science. 2020. PMID: 33184237 Free PMC article.
-
A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming.Science. 2020 Dec 18;370(6523):eabc9546. doi: 10.1126/science.abc9546. Epub 2020 Nov 12. Science. 2020. PMID: 33184234 Free PMC article.
-
Target-directed microRNA degradation: Mechanisms, significance, and functional implications.Wiley Interdiscip Rev RNA. 2024 Mar-Apr;15(2):e1832. doi: 10.1002/wrna.1832. Wiley Interdiscip Rev RNA. 2024. PMID: 38448799 Free PMC article. Review.
Cited by
-
Screening of Drosophila microRNA-degradation sequences reveals Argonaute1 mRNA's role in regulating miR-999.Nat Commun. 2023 Apr 13;14(1):2108. doi: 10.1038/s41467-023-37819-9. Nat Commun. 2023. PMID: 37055443 Free PMC article.
-
Influence of RNA circularity on Target RNA-Directed MicroRNA Degradation.Nucleic Acids Res. 2024 Apr 12;52(6):3358-3374. doi: 10.1093/nar/gkae094. Nucleic Acids Res. 2024. PMID: 38381063 Free PMC article.
-
ZSWIM8 destabilizes many murine microRNAs and is required for proper embryonic growth and development.Genome Res. 2023 Sep;33(9):1482-1496. doi: 10.1101/gr.278073.123. Epub 2023 Aug 2. Genome Res. 2023. PMID: 37532519 Free PMC article.
-
Recent progress in miRNA biogenesis and decay.RNA Biol. 2024 Jan;21(1):1-8. doi: 10.1080/15476286.2023.2288741. Epub 2023 Nov 29. RNA Biol. 2024. PMID: 38031325 Free PMC article. Review.
-
MicroRNA turnover: a tale of tailing, trimming, and targets.Trends Biochem Sci. 2023 Jan;48(1):26-39. doi: 10.1016/j.tibs.2022.06.005. Epub 2022 Jul 7. Trends Biochem Sci. 2023. PMID: 35811249 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
