hnRNPU in Sertoli cells cooperates with WT1 and is essential for testicular development by modulating transcriptional factors Sox8/9
- PMID: 34815802
- PMCID: PMC8581416
- DOI: 10.7150/thno.66819
hnRNPU in Sertoli cells cooperates with WT1 and is essential for testicular development by modulating transcriptional factors Sox8/9
Abstract
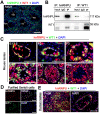
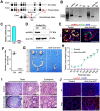
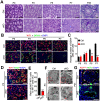
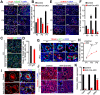
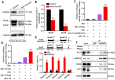
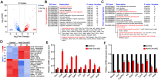
Background: Sertoli cells are essential regulators of testicular fate in the differentiating gonad; however, its role and underlying molecular mechanism of regulating testicular development in prepubertal testes are poorly understood. Although several critical regulatory factors of Sertoli cell development and function have been identified, identifying extrinsic factors that regulate gonocyte proliferation and migration processes during neonatal testis development remains largely unknown. Methods: We used the Sertoli cell-specific conditional knockout strategy (Cre/Loxp) in mice and molecular biological analyses (Luciferase assay, ChIP-qPCR, RNA-Seq, etc.) in vitro and in vivo to study the physiological roles of hnRNPU in Sertoli cells on regulating testicular development in prepubertal testes. Results: We identified a co-transcription factor, hnRNPU, which is highly expressed in mouse and human Sertoli cells and required for neonatal Sertoli cell and pre-pubertal testicular development. Conditional knockout of hnRNPU in murine Sertoli cells leads to severe testicular atrophy and male sterility, characterized by rapid depletion of both Sertoli cells and germ cells and failure of spermatogonia proliferation and migration during pre-pubertal testicular development. At molecular levels, we found that hnRNPU interacts with two Sertoli cell markers WT1 and SOX9, and enhances the expression of two transcriptional factors, Sox8 and Sox9, in Sertoli cells by directly binding to their promoter regions. Further RNA-Seq and bioinformatics analyses revealed the transcriptome-wide of key genes essential for Sertoli cell and germ cell fate control, such as biological adhesion, proliferation and migration, were deregulated in Sertoli cell-specific hnRNPU mutant testes. Conclusion: Our findings demonstrate an essential role of hnRNPU in Sertoli cells for prepubertal testicular development and testis microenvironment maintenance and define a new insight for our understanding of male infertility therapy.
Keywords: Mice; Sertoli cells; Testicular development; Transcription factor; hnRNPU.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
Similar articles
-
Genome-wide identification of Sox8-, and Sox9-dependent genes during early post-natal testis development in the mouse.Andrology. 2013 Mar;1(2):281-92. doi: 10.1111/j.2047-2927.2012.00049.x. Epub 2013 Jan 13. Andrology. 2013. PMID: 23315995
-
Sox9 and Sox8 are required for basal lamina integrity of testis cords and for suppression of FOXL2 during embryonic testis development in mice.Biol Reprod. 2012 Oct 25;87(4):99. doi: 10.1095/biolreprod.112.101907. Print 2012 Oct. Biol Reprod. 2012. PMID: 22837482
-
Testis cord differentiation after the sex determination stage is independent of Sox9 but fails in the combined absence of Sox9 and Sox8.Dev Biol. 2009 Mar 15;327(2):301-12. doi: 10.1016/j.ydbio.2008.12.011. Epub 2008 Dec 24. Dev Biol. 2009. PMID: 19124014
-
SOX E genes: SOX9 and SOX8 in mammalian testis development.Int J Biochem Cell Biol. 2010 Mar;42(3):433-6. doi: 10.1016/j.biocel.2009.07.015. Epub 2009 Jul 30. Int J Biochem Cell Biol. 2010. PMID: 19647095 Review.
-
Role of connexin 43 in Sertoli cells of testis.Ann N Y Acad Sci. 2007 Dec;1120:131-43. doi: 10.1196/annals.1411.004. Epub 2007 Sep 28. Ann N Y Acad Sci. 2007. PMID: 17905936 Review.
Cited by
-
Early Gonadal Development and Sex Determination in Mammal.Int J Mol Sci. 2022 Jul 6;23(14):7500. doi: 10.3390/ijms23147500. Int J Mol Sci. 2022. PMID: 35886859 Free PMC article. Review.
-
Microenvironment of spermatogonial stem cells: a key factor in the regulation of spermatogenesis.Stem Cell Res Ther. 2024 Sep 11;15(1):294. doi: 10.1186/s13287-024-03893-z. Stem Cell Res Ther. 2024. PMID: 39256786 Free PMC article. Review.
-
Development of a novel testis-on-a-chip that demonstrates reciprocal crosstalk between Sertoli and Leydig cells in testicular tissue.Exp Mol Med. 2024 Jul;56(7):1591-1605. doi: 10.1038/s12276-024-01258-3. Epub 2024 Jul 1. Exp Mol Med. 2024. PMID: 38945952 Free PMC article.
-
Effect of Zearalenone-Induced Ferroptosis on Mice Spermatogenesis.Animals (Basel). 2022 Nov 3;12(21):3026. doi: 10.3390/ani12213026. Animals (Basel). 2022. PMID: 36359150 Free PMC article.
-
The single-cell chromatin accessibility landscape in mouse perinatal testis development.Elife. 2023 Apr 25;12:e75624. doi: 10.7554/eLife.75624. Elife. 2023. PMID: 37096870 Free PMC article.
References
-
- Chojnacka K, Zarzycka M, Mruk DD. Biology of the Sertoli Cell in the Fetal, Pubertal, and Adult Mammalian Testis. Results Probl Cell Differ. 2016;58:225–51. - PubMed
-
- Bellve AR. Introduction: the male germ cell; origin, migration, proliferation and differentiation. Semin Cell Dev Biol. 1998;9:379–91. - PubMed
-
- Griswold MD. The central role of Sertoli cells in spermatogenesis. Semin Cell Dev Biol. 1998;9:411–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

