Core genes involved in the regulation of acute lung injury and their association with COVID-19 and tumor progression: A bioinformatics and experimental study
- PMID: 34807957
- PMCID: PMC8608348
- DOI: 10.1371/journal.pone.0260450
Core genes involved in the regulation of acute lung injury and their association with COVID-19 and tumor progression: A bioinformatics and experimental study
Abstract
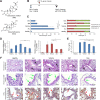
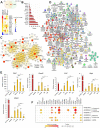
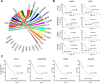
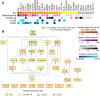
Acute lung injury (ALI) is a specific form of lung damage caused by different infectious and non-infectious agents, including SARS-CoV-2, leading to severe respiratory and systemic inflammation. To gain deeper insight into the molecular mechanisms behind ALI and to identify core elements of the regulatory network associated with this pathology, key genes involved in the regulation of the acute lung inflammatory response (Il6, Ccl2, Cat, Serpine1, Eln, Timp1, Ptx3, Socs3) were revealed using comprehensive bioinformatics analysis of whole-genome microarray datasets, functional annotation of differentially expressed genes (DEGs), reconstruction of protein-protein interaction networks and text mining. The bioinformatics data were validated using a murine model of LPS-induced ALI; changes in the gene expression patterns were assessed during ALI progression and prevention by anti-inflammatory therapy with dexamethasone and the semisynthetic triterpenoid soloxolone methyl (SM), two agents with different mechanisms of action. Analysis showed that 7 of 8 revealed ALI-related genes were susceptible to LPS challenge (up-regulation: Il6, Ccl2, Cat, Serpine1, Eln, Timp1, Socs3; down-regulation: Cat) and their expression was reversed by the pre-treatment of mice with both anti-inflammatory agents. Furthermore, ALI-associated nodal genes were analysed with respect to SARS-CoV-2 infection and lung cancers. The overlap with DEGs identified in postmortem lung tissues from COVID-19 patients revealed genes (Saa1, Rsad2, Ifi44, Rtp4, Mmp8) that (a) showed a high degree centrality in the COVID-19-related regulatory network, (b) were up-regulated in murine lungs after LPS administration, and (c) were susceptible to anti-inflammatory therapy. Analysis of ALI-associated key genes using The Cancer Genome Atlas showed their correlation with poor survival in patients with lung neoplasias (Ptx3, Timp1, Serpine1, Plaur). Taken together, a number of key genes playing a core function in the regulation of lung inflammation were found, which can serve both as promising therapeutic targets and molecular markers to control lung ailments, including COVID-19-associated ALI.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Identification of biomarkers and candidate small-molecule drugs in lipopolysaccharide (LPS)-induced acute lung injury by bioinformatics analysis.Allergol Immunopathol (Madr). 2023 Jan 1;51(1):44-53. doi: 10.15586/aei.v51i1.755. eCollection 2023. Allergol Immunopathol (Madr). 2023. PMID: 36617821
-
An integrated network pharmacology and RNA-Seq approach for exploring the preventive effect of Lonicerae japonicae flos on LPS-induced acute lung injury.J Ethnopharmacol. 2021 Jan 10;264:113364. doi: 10.1016/j.jep.2020.113364. Epub 2020 Sep 9. J Ethnopharmacol. 2021. PMID: 32916233 Free PMC article.
-
Gypenosides Inhibit Inflammatory Response and Apoptosis of Endothelial and Epithelial Cells in LPS-Induced ALI: A Study Based on Bioinformatic Analysis and in vivo/vitro Experiments.Drug Des Devel Ther. 2021 Jan 25;15:289-303. doi: 10.2147/DDDT.S286297. eCollection 2021. Drug Des Devel Ther. 2021. PMID: 33531796 Free PMC article.
-
Delivery systems of therapeutic nucleic acids for the treatment of acute lung injury/acute respiratory distress syndrome.J Control Release. 2023 Aug;360:1-14. doi: 10.1016/j.jconrel.2023.06.018. Epub 2023 Jun 19. J Control Release. 2023. PMID: 37330013 Review.
-
Science review: searching for gene candidates in acute lung injury.Crit Care. 2004 Dec;8(6):440-7. doi: 10.1186/cc2901. Epub 2004 Jun 30. Crit Care. 2004. PMID: 15566614 Free PMC article. Review.
Cited by
-
Classification and severity progression measure of COVID-19 patients using pairs of multi-omic factors.J Appl Stat. 2022 May 4;50(11-12):2473-2503. doi: 10.1080/02664763.2022.2064975. eCollection 2023. J Appl Stat. 2022. PMID: 37529561 Free PMC article.
-
Clinical and CSF single-cell profiling of post-COVID-19 cognitive impairment.Cell Rep Med. 2024 May 21;5(5):101561. doi: 10.1016/j.xcrm.2024.101561. Epub 2024 May 13. Cell Rep Med. 2024. PMID: 38744274 Free PMC article.
-
siRNA-Mediated Timp1 Silencing Inhibited the Inflammatory Phenotype during Acute Lung Injury.Int J Mol Sci. 2023 Jan 13;24(2):1641. doi: 10.3390/ijms24021641. Int J Mol Sci. 2023. PMID: 36675165 Free PMC article.
-
Novel Epoxides of Soloxolone Methyl: An Effect of the Formation of Oxirane Ring and Stereoisomerism on Cytotoxic Profile, Anti-Metastatic and Anti-Inflammatory Activities In Vitro and In Vivo.Int J Mol Sci. 2022 Jun 1;23(11):6214. doi: 10.3390/ijms23116214. Int J Mol Sci. 2022. PMID: 35682893 Free PMC article.
-
Single-cell RNA sequencing reveals hub genes of myocardial infarction-associated endothelial cells.BMC Cardiovasc Disord. 2024 Jan 24;24(1):70. doi: 10.1186/s12872-024-03727-z. BMC Cardiovasc Disord. 2024. PMID: 38267885 Free PMC article.
References
-
- Lee JW, Chun W, Kwon OK, Park HA, Lim Y, Lee JH, et al.. 3,4,5-Trihydroxycinnamic acid attenuates lipopolysaccharide (LPS)-induced acute lung injury via downregulating inflammatory molecules and upregulating HO-1/AMPK activation. Int Immunopharmacol. 2018;64: 123–130. doi: 10.1016/j.intimp.2018.08.015 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

