Ecology, evolution and spillover of coronaviruses from bats
- PMID: 34799704
- PMCID: PMC8603903
- DOI: 10.1038/s41579-021-00652-2
Ecology, evolution and spillover of coronaviruses from bats
Erratum in
-
Author Correction: Ecology, evolution and spillover of coronaviruses from bats.Nat Rev Microbiol. 2022 May;20(5):315. doi: 10.1038/s41579-022-00691-3. Nat Rev Microbiol. 2022. PMID: 35027705 Free PMC article. No abstract available.
Abstract
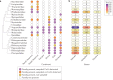
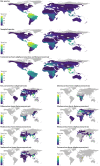
In the past two decades, three coronaviruses with ancestral origins in bats have emerged and caused widespread outbreaks in humans, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Since the first SARS epidemic in 2002-2003, the appreciation of bats as key hosts of zoonotic coronaviruses has advanced rapidly. More than 4,000 coronavirus sequences from 14 bat families have been identified, yet the true diversity of bat coronaviruses is probably much greater. Given that bats are the likely evolutionary source for several human coronaviruses, including strains that cause mild upper respiratory tract disease, their role in historic and future pandemics requires ongoing investigation. We review and integrate information on bat-coronavirus interactions at the molecular, tissue, host and population levels. We identify critical gaps in knowledge of bat coronaviruses, which relate to spillover and pandemic risk, including the pathways to zoonotic spillover, the infection dynamics within bat reservoir hosts, the role of prior adaptation in intermediate hosts for zoonotic transmission and the viral genotypes or traits that predict zoonotic capacity and pandemic potential. Filling these knowledge gaps may help prevent the next pandemic.
© 2021. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Epidemiology and Genomic Characterization of Two Novel SARS-Related Coronaviruses in Horseshoe Bats from Guangdong, China.mBio. 2022 Jun 28;13(3):e0046322. doi: 10.1128/mbio.00463-22. Epub 2022 Apr 25. mBio. 2022. PMID: 35467426 Free PMC article.
-
The Role of Molossidae and Vespertilionidae in Shaping the Diversity of Alphacoronaviruses in the Americas.Microbiol Spectr. 2022 Dec 21;10(6):e0314322. doi: 10.1128/spectrum.03143-22. Epub 2022 Oct 12. Microbiol Spectr. 2022. PMID: 36222689 Free PMC article.
-
[Source of the COVID-19 pandemic: ecology and genetics of coronaviruses (Betacoronavirus: Coronaviridae) SARS-CoV, SARS-CoV-2 (subgenus Sarbecovirus), and MERS-CoV (subgenus Merbecovirus).].Vopr Virusol. 2020;65(2):62-70. doi: 10.36233/0507-4088-2020-65-2-62-70. Vopr Virusol. 2020. PMID: 32515561 Review. Russian.
-
Spike-Independent Infection of Human Coronavirus 229E in Bat Cells.Microbiol Spectr. 2023 Jun 15;11(3):e0348322. doi: 10.1128/spectrum.03483-22. Epub 2023 May 18. Microbiol Spectr. 2023. PMID: 37199653 Free PMC article.
-
Coronaviruses in humans and animals: the role of bats in viral evolution.Environ Sci Pollut Res Int. 2021 Apr;28(16):19589-19600. doi: 10.1007/s11356-021-12553-1. Epub 2021 Mar 2. Environ Sci Pollut Res Int. 2021. PMID: 33655480 Free PMC article. Review.
Cited by
-
Presence of coronaviruses in the common pipistrelle (P. pipistrellus) and Nathusius´ pipistrelle (P. nathusii) in relation to landscape composition.PLoS One. 2023 Nov 29;18(11):e0293649. doi: 10.1371/journal.pone.0293649. eCollection 2023. PLoS One. 2023. PMID: 38019737 Free PMC article.
-
Functional and antigenic characterization of SARS-CoV-2 spike fusion peptide by deep mutational scanning.bioRxiv [Preprint]. 2023 Nov 29:2023.11.28.569051. doi: 10.1101/2023.11.28.569051. bioRxiv. 2023. Update in: Nat Commun. 2024 May 14;15(1):4056. doi: 10.1038/s41467-024-48104-8. PMID: 38076875 Free PMC article. Updated. Preprint.
-
Crystal Structures of Fusion Cores from CCoV-HuPn-2018 and SADS-CoV.Viruses. 2024 Feb 9;16(2):272. doi: 10.3390/v16020272. Viruses. 2024. PMID: 38400047 Free PMC article.
-
Conserved structures and dynamics in 5'-proximal regions of Betacoronavirus RNA genomes.Nucleic Acids Res. 2024 Apr 12;52(6):3419-3432. doi: 10.1093/nar/gkae144. Nucleic Acids Res. 2024. PMID: 38426934 Free PMC article.
-
Bats-associated beta-coronavirus detection and characterization: First report from Pakistan.Infect Genet Evol. 2023 Mar;108:105399. doi: 10.1016/j.meegid.2022.105399. Epub 2022 Dec 27. Infect Genet Evol. 2023. PMID: 36584905 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

