Deubiquitination of proteasome subunits by OTULIN regulates type I IFN production
- PMID: 34797715
- PMCID: PMC8604410
- DOI: 10.1126/sciadv.abi6794
Deubiquitination of proteasome subunits by OTULIN regulates type I IFN production
Abstract
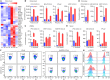
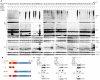
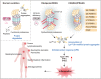
OTULIN is a linear deubiquitinase that negatively regulates the nuclear factor κB (NF-κB) signaling pathway. Patients with OTULIN deficiency, termed as otulipenia or OTULIN-related autoinflammatory syndrome, present with early onset severe systemic inflammation due to increased NF-κB activation. We aimed to investigate additional disease mechanisms of OTULIN deficiency. Our study found a remarkable activation of type I interferon (IFN-I) signaling in whole blood, peripheral blood mononuclear cells, monocytes, and serum from patients with OTULIN deficiency. We observed similar immunologic findings in OTULIN-deficient cell lines generated by CRISPR. Mechanistically, we identified proteasome subunits as substrates of OTULIN deubiquitinase activity and demonstrated proteasome dysregulation in OTULIN-deficient cells as the cause of IFN-I activation. These results reveal an important role of linear ubiquitination in the regulation of proteasome function and suggest a link in the pathogenesis of proteasome-associated autoinflammatory syndromes and OTULIN deficiency.
Figures

Similar articles
-
The Superimposed Deubiquitination Effect of OTULIN and Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Nsp11 Promotes Multiplication of PRRSV.J Virol. 2018 Apr 13;92(9):e00175-18. doi: 10.1128/JVI.00175-18. Print 2018 May 1. J Virol. 2018. PMID: 29444948 Free PMC article.
-
NF-κB Pathway in Autoinflammatory Diseases: Dysregulation of Protein Modifications by Ubiquitin Defines a New Category of Autoinflammatory Diseases.Front Immunol. 2017 Apr 19;8:399. doi: 10.3389/fimmu.2017.00399. eCollection 2017. Front Immunol. 2017. PMID: 28469620 Free PMC article. Review.
-
Reciprocal interplay between OTULIN-LUBAC determines genotoxic and inflammatory NF-κB signal responses.Proc Natl Acad Sci U S A. 2022 Aug 16;119(33):e2123097119. doi: 10.1073/pnas.2123097119. Epub 2022 Aug 8. Proc Natl Acad Sci U S A. 2022. PMID: 35939695 Free PMC article.
-
Biallelic hypomorphic mutations in a linear deubiquitinase define otulipenia, an early-onset autoinflammatory disease.Proc Natl Acad Sci U S A. 2016 Sep 6;113(36):10127-32. doi: 10.1073/pnas.1612594113. Epub 2016 Aug 24. Proc Natl Acad Sci U S A. 2016. PMID: 27559085 Free PMC article.
-
New data in causes of autoinflammatory diseases.Joint Bone Spine. 2019 Oct;86(5):554-561. doi: 10.1016/j.jbspin.2018.11.003. Epub 2018 Nov 22. Joint Bone Spine. 2019. PMID: 30471422 Review.
Cited by
-
OTULIN-related conditions: Report of a new case and review of the literature using GenIA.Res Sq [Preprint]. 2024 Mar 8:rs.3.rs-3950863. doi: 10.21203/rs.3.rs-3950863/v2. Res Sq. 2024. Update in: Clin Immunol. 2024 Aug;265:110292. doi: 10.1016/j.clim.2024.110292. PMID: 38712244 Free PMC article. Updated. Preprint.
-
Regulation of cGAS/STING signaling and corresponding immune escape strategies of viruses.Front Cell Infect Microbiol. 2022 Sep 14;12:954581. doi: 10.3389/fcimb.2022.954581. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36189363 Free PMC article. Review.
-
Disorders of ubiquitylation: unchained inflammation.Nat Rev Rheumatol. 2022 Aug;18(8):435-447. doi: 10.1038/s41584-022-00778-4. Epub 2022 May 6. Nat Rev Rheumatol. 2022. PMID: 35523963 Free PMC article. Review.
-
OTULIN haploinsufficiency predisposes to environmentally directed inflammation.Front Immunol. 2024 May 21;15:983686. doi: 10.3389/fimmu.2024.983686. eCollection 2024. Front Immunol. 2024. PMID: 38827742 Free PMC article.
-
A gain-of-function variation in PLCG1 causes a new immune dysregulation disease.J Allergy Clin Immunol. 2023 Nov;152(5):1292-1302. doi: 10.1016/j.jaci.2023.06.020. Epub 2023 Jul 6. J Allergy Clin Immunol. 2023. PMID: 37422272 Free PMC article.
References
-
- Schaeffer V., Akutsu M., Olma M. H., Gomes L. C., Kawasaki M., Dikic I., Binding of OTULIN to the PUB domain of HOIP controls NF-κB signaling. Mol. Cell 54, 349–361 (2014). - PubMed
-
- Rivkin E., Almeida S. M., Ceccarelli D. F., Juang Y.-C., MacLean T. A., Srikumar T., Huang H., Dunham W. H., Fukumura R., Xie G., Gondo Y., Raught B., Gingras A.-C., Sicheri F., Cordes S. P., The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature 498, 318–324 (2013). - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

