Dysmyelination by Oligodendrocyte-Specific Ablation of Ninj2 Contributes to Depressive-Like Behaviors
- PMID: 34787377
- PMCID: PMC8787401
- DOI: 10.1002/advs.202103065
Dysmyelination by Oligodendrocyte-Specific Ablation of Ninj2 Contributes to Depressive-Like Behaviors
Abstract
Depression is a mental disorder affecting more than 300 million people in the world. Abnormalities in white matter are associated with the development of depression. Here, the authors show that mice with oligodendrocyte-specific deletion of Nerve injury-induced protein 2 (Ninj2) exhibit depressive-like behaviors. Loss of Ninj2 in oligodendrocytes inhibits oligodendrocyte development and myelination, and impairs neuronal structure and activities. Ninj2 competitively inhibits TNFα/TNFR1 signaling pathway by directly binding to TNFR1 in oligodendrocytes. Loss of Ninj2 activates TNFα-induced necroptosis, and increases C-C Motif Chemokine Ligand 2 (Ccl2) production, which might mediate the signal transduction from oligodendrocyte to neurons. Inhibition of necroptosis by Nec-1s administration synchronously restores oligodendrocyte development, improves neuronal excitability, and alleviates depressive-like behaviors. This study thus illustrates the role of Ninj2 in the development of depression and myelination, reveals the relationship between oligodendrocytes and neurons, and provides a potential therapeutic target for depression.
Keywords: Ninj2; depression; necroptosis; oligodendrocytes.
© 2021 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

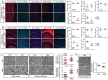

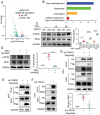
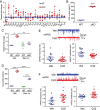
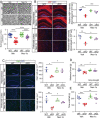

Similar articles
-
Sustained ErbB Activation Causes Demyelination and Hypomyelination by Driving Necroptosis of Mature Oligodendrocytes and Apoptosis of Oligodendrocyte Precursor Cells.J Neurosci. 2021 Dec 1;41(48):9872-9890. doi: 10.1523/JNEUROSCI.2922-20.2021. Epub 2021 Nov 1. J Neurosci. 2021. PMID: 34725188 Free PMC article.
-
Loss of Tuberous Sclerosis Complex1 in Adult Oligodendrocyte Progenitor Cells Enhances Axon Remyelination and Increases Myelin Thickness after a Focal Demyelination.J Neurosci. 2017 Aug 2;37(31):7534-7546. doi: 10.1523/JNEUROSCI.3454-16.2017. Epub 2017 Jul 10. J Neurosci. 2017. PMID: 28694334 Free PMC article.
-
A transgenic mouse model for inducible and reversible dysmyelination.J Neurosci. 2000 Oct 15;20(20):7698-705. doi: 10.1523/JNEUROSCI.20-20-07698.2000. J Neurosci. 2000. PMID: 11027231 Free PMC article.
-
Is psychosis a dysmyelination-related information-processing disorder?Psychiatriki. 2019 Jul-Sep;30(3):245-255. doi: 10.22365/jpsych.2019.303.245. Psychiatriki. 2019. PMID: 31685456 Review.
-
Severe Convulsions and Dysmyelination in Both Jimpy and Cx32/47 -/- Mice may Associate Astrocytic L-Channel Function with Myelination and Oligodendrocytic Connexins with Internodal Kv Channels.Neurochem Res. 2017 Jun;42(6):1747-1766. doi: 10.1007/s11064-017-2194-z. Epub 2017 Feb 18. Neurochem Res. 2017. PMID: 28214987 Review.
Cited by
-
Brain region dependent molecular signatures and myelin repair following chronic demyelination.Front Cell Neurosci. 2023 Apr 26;17:1169786. doi: 10.3389/fncel.2023.1169786. eCollection 2023. Front Cell Neurosci. 2023. PMID: 37180951 Free PMC article.
-
Locus Coeruleus-Dorsolateral Septum Projections Modulate Depression-Like Behaviors via BDNF But Not Norepinephrine.Adv Sci (Weinh). 2024 Mar;11(10):e2303503. doi: 10.1002/advs.202303503. Epub 2023 Dec 28. Adv Sci (Weinh). 2024. PMID: 38155473 Free PMC article.
-
Three-Dimensional Cell Cultures: The Bridge between In Vitro and In Vivo Models.Int J Mol Sci. 2023 Jul 27;24(15):12046. doi: 10.3390/ijms241512046. Int J Mol Sci. 2023. PMID: 37569426 Free PMC article. Review.
-
A new era for myelin research in Neurofibromatosis type 1.Glia. 2023 Dec;71(12):2701-2719. doi: 10.1002/glia.24432. Epub 2023 Jun 29. Glia. 2023. PMID: 37382486 Free PMC article. Review.
-
Neuroimmune Mechanisms Underlying Neuropathic Pain: The Potential Role of TNF-α-Necroptosis Pathway.Int J Mol Sci. 2022 Jun 28;23(13):7191. doi: 10.3390/ijms23137191. Int J Mol Sci. 2022. PMID: 35806192 Free PMC article. Review.
References
-
- Malhi G. S., Mann J. J., Lancet 2018, 392, 2299. - PubMed
-
- a) Shen Z. L., Cheng Y. Q., Yang S. R., Dai N., Ye J., Liu X. Y., Lu J., Li N., Liu F., Lu Y., Sun X. J., Xu X. F., Neuroimage: Clin. 2016, 12, 492; - PMC - PubMed
- b) Ransome M. I., Renoir T., Hannan A. J., Neural Plast. 2012, 2012, 874387; - PMC - PubMed
- c) Zhuo C. J., Zhu J. J., Wang C. L., Qu H. R., Ma X. L., Qin W., Brain Imaging Behav. 2017, 11, 1678. - PMC - PubMed
-
- Baumann N., Pham‐Dinh D., Physiol. Rev. 2001, 81, 871. - PubMed
-
- Nave K. A., Edgar J., Griffiths I. R., Jansen K., Kassmann C. M., Klugmann M., Lappe‐Siefke C., Meyer‐zu‐Horste G., Mobius W., Sereda M. H., Werner H., J. Neuroimmunol. 2006, 178, 36.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
