mTOR-mediated phosphorylation of VAMP8 and SCFD1 regulates autophagosome maturation
- PMID: 34785650
- PMCID: PMC8595342
- DOI: 10.1038/s41467-021-26824-5
mTOR-mediated phosphorylation of VAMP8 and SCFD1 regulates autophagosome maturation
Abstract
The mammalian target of rapamycin (mTORC1) has been shown to regulate autophagy at different steps. However, how mTORC1 regulates the N-ethylmaleimide-sensitive protein receptor (SNARE) complex remains elusive. Here we show that mTORC1 inhibits formation of the SNARE complex (STX17-SNAP29-VAMP8) by phosphorylating VAMP8, thereby blocking autophagosome-lysosome fusion. A VAMP8 phosphorylation mimic mutant is unable to promote autophagosome-lysosome fusion in vitro. Furthermore, we identify SCFD1, a Sec1/Munc18-like protein, that localizes to the autolysosome and is required for SNARE complex formation and autophagosome-lysosome fusion. VAMP8 promotes SCFD1 recruitment to autolysosomes when dephosphorylated. Consistently, phosphorylated VAMP8 or SCFD1 depletion inhibits autophagosome-lysosome fusion, and expression of phosphomimic VAMP8 leads to increased lipid droplet accumulation when expressed in mouse liver. Thus, our study supports that mTORC1-mediated phosphorylation of VAMP8 blocks SCFD1 recruitment, thereby inhibiting STX17-SNAP29-VAMP8 complex formation and autophagosome-lysosome fusion.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

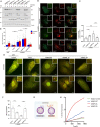
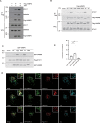
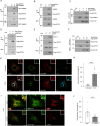
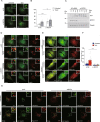

Comment in
-
MTOR-mediates hepatic lipid metabolism through an autophagic SNARE complex.Autophagy. 2022 Jun;18(6):1467-1469. doi: 10.1080/15548627.2022.2037853. Epub 2022 Feb 17. Autophagy. 2022. PMID: 35174767 Free PMC article.
Similar articles
-
MTOR-mediates hepatic lipid metabolism through an autophagic SNARE complex.Autophagy. 2022 Jun;18(6):1467-1469. doi: 10.1080/15548627.2022.2037853. Epub 2022 Feb 17. Autophagy. 2022. PMID: 35174767 Free PMC article.
-
ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes.Nature. 2015 Apr 23;520(7548):563-6. doi: 10.1038/nature14147. Epub 2015 Feb 9. Nature. 2015. PMID: 25686604 Free PMC article.
-
Acetylation of SCFD1 regulates SNARE complex formation and autophagosome-lysosome fusion.Autophagy. 2023 Jan;19(1):189-203. doi: 10.1080/15548627.2022.2064624. Epub 2022 Apr 24. Autophagy. 2023. PMID: 35465820 Free PMC article.
-
New insights regarding SNARE proteins in autophagosome-lysosome fusion.Autophagy. 2021 Oct;17(10):2680-2688. doi: 10.1080/15548627.2020.1823124. Epub 2020 Sep 24. Autophagy. 2021. PMID: 32924745 Free PMC article. Review.
-
The multi-functional SNARE protein Ykt6 in autophagosomal fusion processes.Cell Cycle. 2019 Mar-Apr;18(6-7):639-651. doi: 10.1080/15384101.2019.1580488. Epub 2019 Mar 17. Cell Cycle. 2019. PMID: 30836834 Free PMC article. Review.
Cited by
-
Investigating Causality and Shared Genetic Architecture between Neurodegenerative Disorders and Inflammatory Bowel Disease.Aging Dis. 2023 Aug 1;14(4):1349-1359. doi: 10.14336/AD.2022.12209. Aging Dis. 2023. PMID: 37163440 Free PMC article.
-
Molecular Mechanism of Autophagosome-Lysosome Fusion in Mammalian Cells.Cells. 2024 Mar 13;13(6):500. doi: 10.3390/cells13060500. Cells. 2024. PMID: 38534345 Free PMC article. Review.
-
mTOR Deletion Alleviates CD4+ T-Cell Dysfunction in Sepsis through Reducing CTLA4 Accumulation Mediated by Rescuing Autophagy.Mediators Inflamm. 2024 Jul 25;2024:4233439. doi: 10.1155/2024/4233439. eCollection 2024. Mediators Inflamm. 2024. PMID: 39104632 Free PMC article.
-
Accumulation of Intracellular Protein Advanced Glycation End Products Alters Cellular Homeostasis for Protein Clearance in Pancreatic Ductal Epithelial Cells.Biochemistry. 2024 Jul 16;63(14):1723-1729. doi: 10.1021/acs.biochem.4c00250. Epub 2024 Jun 28. Biochemistry. 2024. PMID: 38941592
-
Axonopathy Underlying Amyotrophic Lateral Sclerosis: Unraveling Complex Pathways and Therapeutic Insights.Neurosci Bull. 2024 Nov;40(11):1789-1810. doi: 10.1007/s12264-024-01267-2. Epub 2024 Aug 4. Neurosci Bull. 2024. PMID: 39097850 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

