New insights into Epstein‑Barr virus‑associated tumors: Exosomes (Review)
- PMID: 34779497
- PMCID: PMC8600424
- DOI: 10.3892/or.2021.8224
New insights into Epstein‑Barr virus‑associated tumors: Exosomes (Review)
Abstract
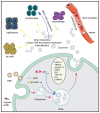
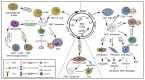
Epstein‑Barr virus (EBV) is endemic worldwide and is associated with a number of human tumors. EBV‑associated tumors have unique mechanisms of tumorigenesis. EBV encodes multiple oncogenic molecules that can be loaded into exosomes released by EBV+ tumor cells to mediate intercellular communication. Moreover, different EBV+ tumor cells secrete exosomes that act on various target cells with various biological functions. In addition to oncogenicity, EBV+ exosomes have potential immunosuppressive effects. Investigating EBV+ exosomes could identify the role of EBV in tumorigenesis and progression. The present review summarized advances in studies focusing on exosomes and the functions of EBV+ exosomes derived from different EBV‑associated tumors. EBV+ exosomes are expected to become a new biomarker for disease diagnosis and prognosis. Therefore, exosome‑targeted therapy displays potential.
Keywords: Epstein‑Barr virus; biomarker; exosome; immunosuppressive; microRNAs.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Modulation of B-cell exosome proteins by gamma herpesvirus infection.Proc Natl Acad Sci U S A. 2013 Jul 30;110(31):E2925-33. doi: 10.1073/pnas.1303906110. Epub 2013 Jul 1. Proc Natl Acad Sci U S A. 2013. PMID: 23818640 Free PMC article.
-
Pathogenic Role of Exosomes in Epstein-Barr Virus (EBV)-Associated Cancers.Int J Biol Sci. 2017 Sep 21;13(10):1276-1286. doi: 10.7150/ijbs.19531. eCollection 2017. Int J Biol Sci. 2017. PMID: 29104494 Free PMC article. Review.
-
Exosomal cyclophilin A as a novel noninvasive biomarker for Epstein-Barr virus associated nasopharyngeal carcinoma.Cancer Med. 2019 Jun;8(6):3142-3151. doi: 10.1002/cam4.2185. Epub 2019 May 7. Cancer Med. 2019. PMID: 31063269 Free PMC article.
-
Methodological Approaches to Study Extracellular Vesicle miRNAs in Epstein⁻Barr Virus-Associated Cancers.Int J Mol Sci. 2018 Sep 18;19(9):2810. doi: 10.3390/ijms19092810. Int J Mol Sci. 2018. PMID: 30231493 Free PMC article. Review.
-
Exosomal communication goes viral.J Virol. 2015 May;89(10):5200-3. doi: 10.1128/JVI.02470-14. Epub 2015 Mar 4. J Virol. 2015. PMID: 25740980 Free PMC article. Review.
Cited by
-
EBV-microRNAs as Potential Biomarkers in EBV-related Fever: A Narrative Review.Curr Mol Med. 2024;24(1):2-13. doi: 10.2174/1566524023666221118122005. Curr Mol Med. 2024. PMID: 36411555 Free PMC article. Review.
-
Exosomal transmission of viruses, a two-edged biological sword.Cell Commun Signal. 2023 Jan 23;21(1):19. doi: 10.1186/s12964-022-01037-5. Cell Commun Signal. 2023. PMID: 36691072 Free PMC article. Review.
-
Extracellular vesicles and endothelial dysfunction in infectious diseases.J Extracell Biol. 2024 Apr 12;3(4):e148. doi: 10.1002/jex2.148. eCollection 2024 Apr. J Extracell Biol. 2024. PMID: 38938849 Free PMC article. Review.
-
Immunosuppressive Tumor Microenvironment and Immunotherapy of Epstein-Barr Virus-Associated Malignancies.Viruses. 2022 May 10;14(5):1017. doi: 10.3390/v14051017. Viruses. 2022. PMID: 35632758 Free PMC article. Review.
-
Suppression of Nasopharyngeal and Gastric Tumor Growth in a Mouse Model by Antibodies to Epstein-Barr Virus LMP1 Protein.Microorganisms. 2023 Jun 30;11(7):1712. doi: 10.3390/microorganisms11071712. Microorganisms. 2023. PMID: 37512884 Free PMC article.
References
-
- Lyu X, Wang J, Guo X, Wu G, Jiao Y, Faleti OD, Liu P, Liu T, Long Y, Chong T, et al. EBV-miR-BART1-5P activates AMPK/mTOR/HIF1 pathway via a PTEN independent manner to promote glycolysis and angiogenesis in nasopharyngeal carcinoma. PLoS Pathog. 2018;14:e1007484. doi: 10.1371/journal.ppat.1007484. - DOI - PMC - PubMed

