KRAS mutation: from undruggable to druggable in cancer
- PMID: 34776511
- PMCID: PMC8591115
- DOI: 10.1038/s41392-021-00780-4
KRAS mutation: from undruggable to druggable in cancer
Abstract
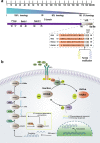
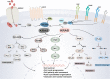
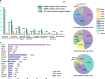
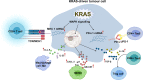
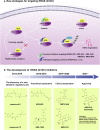
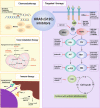
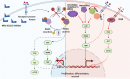
Cancer is the leading cause of death worldwide, and its treatment and outcomes have been dramatically revolutionised by targeted therapies. As the most frequently mutated oncogene, Kirsten rat sarcoma viral oncogene homologue (KRAS) has attracted substantial attention. The understanding of KRAS is constantly being updated by numerous studies on KRAS in the initiation and progression of cancer diseases. However, KRAS has been deemed a challenging therapeutic target, even "undruggable", after drug-targeting efforts over the past four decades. Recently, there have been surprising advances in directly targeted drugs for KRAS, especially in KRAS (G12C) inhibitors, such as AMG510 (sotorasib) and MRTX849 (adagrasib), which have obtained encouraging results in clinical trials. Excitingly, AMG510 was the first drug-targeting KRAS (G12C) to be approved for clinical use this year. This review summarises the most recent understanding of fundamental aspects of KRAS, the relationship between the KRAS mutations and tumour immune evasion, and new progress in targeting KRAS, particularly KRAS (G12C). Moreover, the possible mechanisms of resistance to KRAS (G12C) inhibitors and possible combination therapies are summarised, with a view to providing the best regimen for individualised treatment with KRAS (G12C) inhibitors and achieving truly precise treatment.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Structural insights into small-molecule KRAS inhibitors for targeting KRAS mutant cancers.Eur J Med Chem. 2024 Nov 5;277:116771. doi: 10.1016/j.ejmech.2024.116771. Epub 2024 Aug 15. Eur J Med Chem. 2024. PMID: 39167893 Review.
-
The next-generation KRAS inhibitors…What comes after sotorasib and adagrasib?Lung Cancer. 2024 Aug;194:107886. doi: 10.1016/j.lungcan.2024.107886. Epub 2024 Jul 10. Lung Cancer. 2024. PMID: 39047616 Review.
-
KRAS: From undruggable to a druggable Cancer Target.Cancer Treat Rev. 2020 Sep;89:102070. doi: 10.1016/j.ctrv.2020.102070. Epub 2020 Jul 15. Cancer Treat Rev. 2020. PMID: 32711246 Review.
-
KRAS G12C Game of Thrones, which direct KRAS inhibitor will claim the iron throne?Cancer Treat Rev. 2020 Mar;84:101974. doi: 10.1016/j.ctrv.2020.101974. Epub 2020 Jan 23. Cancer Treat Rev. 2020. PMID: 32014824 Free PMC article. Review.
-
Targeted Therapies for Previously "Undruggable" KRAS-Mutated Non-Small Cell Lung Cancer: A Review of Sotorasib and Adagrasib.Ann Pharmacother. 2024 Jun;58(6):622-635. doi: 10.1177/10600280231197459. Epub 2023 Sep 12. Ann Pharmacother. 2024. PMID: 37700573 Review.
Cited by
-
Mechanistic insights into the clinical Y96D mutation with acquired resistance to AMG510 in the KRASG12C.Front Oncol. 2022 Aug 10;12:915512. doi: 10.3389/fonc.2022.915512. eCollection 2022. Front Oncol. 2022. PMID: 36033504 Free PMC article.
-
Pancreatic STAT5 activation promotes KrasG12D-induced and inflammation-induced acinar-to-ductal metaplasia and pancreatic cancer.Gut. 2024 Oct 7;73(11):1831-1843. doi: 10.1136/gutjnl-2024-332225. Gut. 2024. PMID: 38955401 Free PMC article.
-
A large-scale cancer-specific protein-DNA interaction network.bioRxiv [Preprint]. 2024 Jan 29:2024.01.24.577099. doi: 10.1101/2024.01.24.577099. bioRxiv. 2024. Update in: Life Sci Alliance. 2024 Jul 16;7(10):e202402641. doi: 10.26508/lsa.202402641 PMID: 38352498 Free PMC article. Updated. Preprint.
-
KRAS silencing alters chromatin physical organization and transcriptional activity in colorectal cancer cells.Res Sq [Preprint]. 2024 Apr 16:rs.3.rs-3752760. doi: 10.21203/rs.3.rs-3752760/v2. Res Sq. 2024. PMID: 38410476 Free PMC article. Preprint.
-
Small RNA-Seq Reveals Similar miRNA Transcriptome in Children and Young Adults with T-ALL and Indicates miR-143-3p as Novel Candidate Tumor Suppressor in This Leukemia.Int J Mol Sci. 2022 Sep 4;23(17):10117. doi: 10.3390/ijms231710117. Int J Mol Sci. 2022. PMID: 36077521 Free PMC article.
References
-
- Sequist LV, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J. Clin. Oncol. 2013;31:3327–3334. - PubMed
-
- Mok TS, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009;361:947–957. - PubMed
-
- Soria J-C, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018;378:113–125. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

