Deciphering the Biological Significance of ADAR1-Z-RNA Interactions
- PMID: 34768866
- PMCID: PMC8584189
- DOI: 10.3390/ijms222111435
Deciphering the Biological Significance of ADAR1-Z-RNA Interactions
Abstract
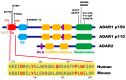
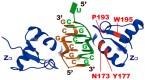
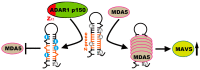
Adenosine deaminase acting on RNA 1 (ADAR1) is an enzyme responsible for double-stranded RNA (dsRNA)-specific adenosine-to-inosine RNA editing, which is estimated to occur at over 100 million sites in humans. ADAR1 is composed of two isoforms transcribed from different promoters: p150 and N-terminal truncated p110. Deletion of ADAR1 p150 in mice activates melanoma differentiation-associated protein 5 (MDA5)-sensing pathway, which recognizes endogenous unedited RNA as non-self. In contrast, we have recently demonstrated that ADAR1 p110-mediated RNA editing does not contribute to this function, implying that a unique Z-DNA/RNA-binding domain α (Zα) in the N terminus of ADAR1 p150 provides specific RNA editing, which is critical for preventing MDA5 activation. In addition, a mutation in the Zα domain is identified in patients with Aicardi-Goutières syndrome (AGS), an inherited encephalopathy characterized by overproduction of type I interferon. Accordingly, we and other groups have recently demonstrated that Adar1 Zα-mutated mice show MDA5-dependent type I interferon responses. Furthermore, one such mutant mouse carrying a W197A point mutation in the Zα domain, which inhibits Z-RNA binding, manifests AGS-like encephalopathy. These findings collectively suggest that Z-RNA binding by ADAR1 p150 is essential for proper RNA editing at certain sites, preventing aberrant MDA5 activation.
Keywords: ADAR1; AGS; IGS; MDA5; RNA editing; Z-RNA; interferonopathy.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


Similar articles
-
The RNA-editing enzyme ADAR1: a regulatory hub that tunes multiple dsRNA-sensing pathways.Int Immunol. 2023 Mar 14;35(3):123-133. doi: 10.1093/intimm/dxac056. Int Immunol. 2023. PMID: 36469491
-
Mutations in the adenosine deaminase ADAR1 that prevent endogenous Z-RNA binding induce Aicardi-Goutières-syndrome-like encephalopathy.Immunity. 2021 Sep 14;54(9):1976-1988.e7. doi: 10.1016/j.immuni.2021.08.022. Immunity. 2021. PMID: 34525338
-
ADAR1 interaction with Z-RNA promotes editing of endogenous double-stranded RNA and prevents MDA5-dependent immune activation.Cell Rep. 2021 Aug 10;36(6):109500. doi: 10.1016/j.celrep.2021.109500. Cell Rep. 2021. PMID: 34380029
-
An RNA editor, adenosine deaminase acting on double-stranded RNA (ADAR1).J Interferon Cytokine Res. 2014 Jun;34(6):437-46. doi: 10.1089/jir.2014.0001. J Interferon Cytokine Res. 2014. PMID: 24905200 Free PMC article. Review.
-
ADAR1, inosine and the immune sensing system: distinguishing self from non-self.Wiley Interdiscip Rev RNA. 2016 Mar-Apr;7(2):157-72. doi: 10.1002/wrna.1322. Epub 2015 Dec 21. Wiley Interdiscip Rev RNA. 2016. PMID: 26692549 Review.
Cited by
-
Structural impacts of two disease-linked ADAR1 mutants: a molecular dynamics study.J Comput Aided Mol Des. 2024 Jul 17;38(1):25. doi: 10.1007/s10822-024-00565-1. J Comput Aided Mol Des. 2024. PMID: 39014124
-
ZBP1: A Powerful Innate Immune Sensor and Double-Edged Sword in Host Immunity.Int J Mol Sci. 2022 Sep 6;23(18):10224. doi: 10.3390/ijms231810224. Int J Mol Sci. 2022. PMID: 36142136 Free PMC article. Review.
-
Generation of a new Adar1p150 -/- mouse demonstrates isoform-specific roles in embryonic development and adult homeostasis.RNA. 2023 Sep;29(9):1325-1338. doi: 10.1261/rna.079509.122. Epub 2023 Jun 8. RNA. 2023. PMID: 37290963 Free PMC article.
-
Differential Structural Features of Two Mutant ADAR1p150 Zα Domains Associated with Aicardi-Goutières Syndrome.J Mol Biol. 2023 Apr 15;435(8):168040. doi: 10.1016/j.jmb.2023.168040. Epub 2023 Mar 7. J Mol Biol. 2023. PMID: 36889460 Free PMC article.
-
Zα domain proteins mediate the immune response.Front Immunol. 2023 Sep 12;14:1241694. doi: 10.3389/fimmu.2023.1241694. eCollection 2023. Front Immunol. 2023. PMID: 37771585 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

