The SR Splicing Factors: Providing Perspectives on Their Evolution, Expression, Alternative Splicing, and Function in Populus trichocarpa
- PMID: 34768799
- PMCID: PMC8583155
- DOI: 10.3390/ijms222111369
The SR Splicing Factors: Providing Perspectives on Their Evolution, Expression, Alternative Splicing, and Function in Populus trichocarpa
Abstract
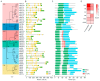
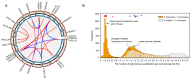
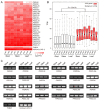
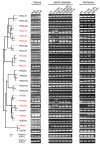
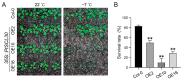
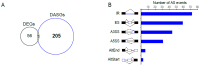
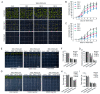
Serine/arginine-rich (SR) proteins are important splicing factors in plant development and abiotic/hormone-related stresses. However, evidence that SR proteins contribute to the process in woody plants has been lacking. Using phylogenetics, gene synteny, transgenic experiments, and RNA-seq analysis, we identified 24 PtSR genes and explored their evolution, expression, and function in Popolus trichocarpa. The PtSR genes were divided into six subfamilies, generated by at least two events of genome triplication and duplication. Notably, they were constitutively expressed in roots, stems, and leaves, demonstrating their fundamental role in P. trichocarpa. Additionally, most PtSR genes (~83%) responded to at least one stress (cold, drought, salt, SA, MeJA, or ABA), and, especially, cold stress induced a dramatic perturbation in the expression and/or alternative splicing (AS) of 18 PtSR genes (~75%). Evidentially, the overexpression of PtSCL30 in Arabidopsis decreased freezing tolerance, which probably resulted from AS changes of the genes (e.g., ICE2 and COR15A) critical for cold tolerance. Moreover, the transgenic plants were salt-hypersensitive at the germination stage. These indicate that PtSCL30 may act as a negative regulator under cold and salt stress. Altogether, this study sheds light on the evolution, expression, and AS of PtSR genes, and the functional mechanisms of PtSCL30 in woody plants.
Keywords: Populus trichocarpa; PtSCL30; abiotic stress; alternative splicing; serine/arginine-rich (SR) protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Systematic analysis of the Serine/Arginine-Rich Protein Splicing Factors (SRs) and focus on salt tolerance of PtSC27 in Populus trichocarpa.Plant Physiol Biochem. 2022 Jan 15;173:97-109. doi: 10.1016/j.plaphy.2022.01.015. Epub 2022 Jan 21. Plant Physiol Biochem. 2022. PMID: 35121529
-
Genome-Wide Comprehensive Analysis of the GASA Gene Family in Populus.Int J Mol Sci. 2021 Nov 15;22(22):12336. doi: 10.3390/ijms222212336. Int J Mol Sci. 2021. PMID: 34830215 Free PMC article.
-
A Stress-Associated Protein, PtSAP13, From Populus trichocarpa Provides Tolerance to Salt Stress.Int J Mol Sci. 2019 Nov 17;20(22):5782. doi: 10.3390/ijms20225782. Int J Mol Sci. 2019. PMID: 31744233 Free PMC article.
-
Functional analyses of NDPK2 in Populus trichocarpa and overexpression of PtNDPK2 enhances growth and tolerance to abiotic stresses in transgenic poplar.Plant Physiol Biochem. 2017 Aug;117:61-74. doi: 10.1016/j.plaphy.2017.05.019. Epub 2017 May 31. Plant Physiol Biochem. 2017. PMID: 28587994
-
A role for SR proteins in plant stress responses.Plant Signal Behav. 2011 Jan;6(1):49-54. doi: 10.4161/psb.6.1.14063. Epub 2011 Jan 1. Plant Signal Behav. 2011. PMID: 21258207 Free PMC article. Review.
Cited by
-
Pan-transcriptomic analysis reveals alternative splicing control of cold tolerance in rice.Plant Cell. 2024 May 29;36(6):2117-2139. doi: 10.1093/plcell/koae039. Plant Cell. 2024. PMID: 38345423
-
Alternative splicing: transcriptional regulatory network in agroforestry.Front Plant Sci. 2023 Apr 12;14:1158965. doi: 10.3389/fpls.2023.1158965. eCollection 2023. Front Plant Sci. 2023. PMID: 37123829 Free PMC article. Review.
-
A phylotranscriptomic dataset of angiosperm species under cold stress.Sci Data. 2023 Jun 22;10(1):399. doi: 10.1038/s41597-023-02307-8. Sci Data. 2023. PMID: 37349352 Free PMC article.
-
Regulatory Network of Serine/Arginine-Rich (SR) Proteins: The Molecular Mechanism and Physiological Function in Plants.Int J Mol Sci. 2022 Sep 5;23(17):10147. doi: 10.3390/ijms231710147. Int J Mol Sci. 2022. PMID: 36077545 Free PMC article. Review.
-
Interplays between cis- and trans-Acting Factors for Alternative Splicing in Response to Environmental Changes during Biological Invasions of Ascidians.Int J Mol Sci. 2023 Oct 5;24(19):14921. doi: 10.3390/ijms241914921. Int J Mol Sci. 2023. PMID: 37834365 Free PMC article.
References
-
- Zhu F.Y., Chen M.X., Ye N.H., Shi L., Ma K.L., Yang J.F., Cao Y.Y., Zhang Y., Yoshida T., Fernie A.R., et al. Proteogenomic analysis reveals alternative splicing and translation as part of the abscisic acid response in Arabidopsis seedlings. Plant J. 2017;91:518–533. doi: 10.1111/tpj.13571. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

