A comprehensive analysis of prefoldins and their implication in cancer
- PMID: 34761191
- PMCID: PMC8567396
- DOI: 10.1016/j.isci.2021.103273
A comprehensive analysis of prefoldins and their implication in cancer
Abstract
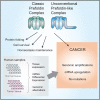
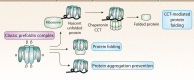
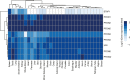
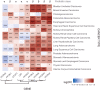
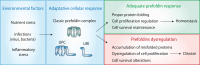
Prefoldins (PFDNs) are evolutionary conserved co-chaperones, initially discovered in archaea but universally present in eukaryotes. PFDNs are prevalently organized into hetero-hexameric complexes. Although they have been overlooked since their discovery and their functions remain elusive, several reports indicate they act as co-chaperones escorting misfolded or non-native proteins to group II chaperonins. Unlike the eukaryotic PFDNs which interact with cytoskeletal components, the archaeal PFDNs can bind and stabilize a wide range of substrates, possibly due to their great structural diversity. The discovery of the unconventional RPB5 interactor (URI) PFDN-like complex (UPC) suggests that PFDNs have versatile functions and are required for different cellular processes, including an important role in cancer. Here, we summarize their functions across different species. Moreover, a comprehensive analysis of PFDNs genomic alterations across cancer types by using large-scale cancer genomic data indicates that PFDNs are a new class of non-mutated proteins significantly overexpressed in some cancer types.
Keywords: Biological sciences; Cancer; Cell biology.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Prefoldin Subunits and Its Associate Partners: Conservations and Specificities in Plants.Plants (Basel). 2024 Feb 18;13(4):556. doi: 10.3390/plants13040556. Plants (Basel). 2024. PMID: 38498526 Free PMC article. Review.
-
Prefoldins in Archaea.Adv Exp Med Biol. 2018;1106:11-23. doi: 10.1007/978-3-030-00737-9_2. Adv Exp Med Biol. 2018. PMID: 30484150 Review.
-
Comprehensive analysis of the prognostic value and functions of prefoldins in hepatocellular carcinoma.Front Mol Biosci. 2022 Nov 11;9:957001. doi: 10.3389/fmolb.2022.957001. eCollection 2022. Front Mol Biosci. 2022. PMID: 36438659 Free PMC article.
-
Role of the Unconventional Prefoldin Proteins URI and UXT in Transcription Regulation.Adv Exp Med Biol. 2018;1106:85-94. doi: 10.1007/978-3-030-00737-9_6. Adv Exp Med Biol. 2018. PMID: 30484154 Review.
-
Evolution of assisted protein folding: the distribution of the main chaperoning systems within the phylogenetic domain archaea.Front Biosci. 2004 May 1;9:1318-32. doi: 10.2741/1328. Front Biosci. 2004. PMID: 14977547 Review.
Cited by
-
Prefoldin Subunits and Its Associate Partners: Conservations and Specificities in Plants.Plants (Basel). 2024 Feb 18;13(4):556. doi: 10.3390/plants13040556. Plants (Basel). 2024. PMID: 38498526 Free PMC article. Review.
-
PFDN2 promotes cell cycle progression via the hnRNPD-MYBL2 axis in gastric cancer.Front Oncol. 2023 Jul 18;13:1164070. doi: 10.3389/fonc.2023.1164070. eCollection 2023. Front Oncol. 2023. PMID: 37538116 Free PMC article.
-
Prefoldin 2 contributes to mitochondrial morphology and function.BMC Biol. 2023 Sep 12;21(1):193. doi: 10.1186/s12915-023-01695-y. BMC Biol. 2023. PMID: 37697385 Free PMC article.
-
Transcriptomics data integration and analysis to uncover hallmark genes in hypertrophic cardiomyopathy.Am J Transl Res. 2024 Feb 15;16(2):637-653. doi: 10.62347/AXOY3338. eCollection 2024. Am J Transl Res. 2024. PMID: 38463581 Free PMC article.
-
Endogenous retroviruses shape pluripotency specification in mouse embryos.Sci Adv. 2024 Jan 26;10(4):eadk9394. doi: 10.1126/sciadv.adk9394. Epub 2024 Jan 24. Sci Adv. 2024. PMID: 38266080 Free PMC article.
References
-
- Ajjappala H., Chung H.Y., Sim J.S., Choi I., Hahn B.S. Disruption of prefoldin-2 protein synthesis in root-knot nematodes via host-mediated gene silencing efficiently reduces nematode numbers and thus protects plants. Planta. 2015;241:773–787. - PubMed
-
- Alldinger I., Dittert D., Peiper M., Fusco A., Chiappetta G., Staub E., Lohr M., Jesnowski R., Baretton G., Ockert D. Gene expression analysis of pancreatic cell lines reveals genes overexpressed in pancreatic cancer. Pancreatology. 2005;5:370–379. - PubMed
-
- Amorim A.F., Pinto D., Kuras L., Fernandes L. Absence of Gim proteins, but not GimC complex, alters stress-induced transcription. Biochim. Biophys. Acta Gene Regul. Mech. 2017;1860:773–781. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

