Alternative Splicing Landscape of Small Brown Planthopper and Different Response of JNK2 Isoforms to Rice Stripe Virus Infection
- PMID: 34757837
- PMCID: PMC8791299
- DOI: 10.1128/JVI.01715-21
Alternative Splicing Landscape of Small Brown Planthopper and Different Response of JNK2 Isoforms to Rice Stripe Virus Infection
Abstract
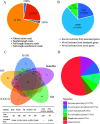
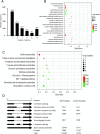
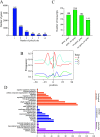
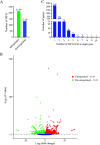
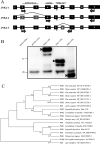
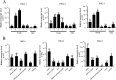
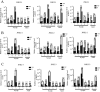
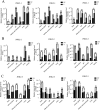
Alternative splicing (AS) is a frequent posttranscriptional regulatory event occurring in response to various endogenous and exogenous stimuli in most eukaryotic organisms. However, little is known about the effects of insect-transmitted viruses on AS events in insect vectors. The present study used third-generation sequencing technology and RNA sequencing (RNA-Seq) to evaluate the AS response in the small brown planthopper Laodelphax striatellus to rice stripe virus (RSV). The full-length transcriptome of L. striatellus was obtained using single-molecule real-time sequencing technology (SMRT). Posttranscriptional regulatory events, including AS, alternative polyadenylation, and fusion transcripts, were analyzed. A total of 28,175 nonredundant transcript isoforms included 24,950 transcripts assigned to 8,500 annotated genes of L. striatellus, and 5,000 of these genes (58.8%) had AS events. RNA-Seq of the gut samples of insects infected by RSV for 8 d identified 3,458 differentially expressed transcripts (DETs); 2,185 of these DETs were transcribed from 1,568 genes that had AS events, indicating that 31.4% of alternatively spliced genes responded to RSV infection of the gut. One of the c-Jun N-terminal kinase (JNK) genes, JNK2, experienced exon skipping, resulting in three transcript isoforms. These three isoforms differentially responded to RSV infection during development and in various organs. Injection of double-stranded RNAs targeting all or two isoforms indicated that three or at least two JNK2 isoforms facilitated RSV accumulation in planthoppers. These results implied that AS events could participate in the regulation of complex relationships between viruses and insect vectors. IMPORTANCE Alternative splicing (AS) is a regulatory mechanism that occurs after gene transcription. AS events can enrich protein diversity to promote the reactions of the organisms to various endogenous and exogenous stimulations. It is not known how insect vectors exploit AS events to cope with transmitted viruses. The present study used third-generation sequencing technology to obtain the profile of AS events in the small brown planthopper Laodelphax striatellus, which is an efficient vector for rice stripe virus (RSV). The results indicated that 31.4% of alternatively spliced genes responded to RSV infection in the gut of planthoppers. One of the c-Jun N-terminal kinase (JNK) genes, JNK2, produced three transcript isoforms by AS. These three isoforms showed different responses to RSV infection, and at least two isoforms facilitated viral accumulation in planthoppers. These results implied that AS events could participate in the regulation of complex relationships between viruses and insect vectors.
Keywords: JNK; RNA-Seq; SMRT; alternative splicing; full-length transcriptome; insect vector; plant virus; rice stripe virus; small brown planthopper.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Rice stripe virus-derived siRNAs play different regulatory roles in rice and in the insect vector Laodelphax striatellus.BMC Plant Biol. 2018 Oct 4;18(1):219. doi: 10.1186/s12870-018-1438-7. BMC Plant Biol. 2018. PMID: 30286719 Free PMC article.
-
Comparative Transcriptome Analysis of Chemoreception Organs of Laodelphax striatellus in Response to Rice Stripe Virus Infection.Int J Mol Sci. 2021 Sep 24;22(19):10299. doi: 10.3390/ijms221910299. Int J Mol Sci. 2021. PMID: 34638638 Free PMC article.
-
Massively parallel pyrosequencing-based transcriptome analyses of small brown planthopper (Laodelphax striatellus), a vector insect transmitting rice stripe virus (RSV).BMC Genomics. 2010 May 13;11:303. doi: 10.1186/1471-2164-11-303. BMC Genomics. 2010. PMID: 20462456 Free PMC article.
-
The small brown planthopper (Laodelphax striatellus) as a vector of the rice stripe virus.Arch Insect Biochem Physiol. 2023 Feb;112(2):e21992. doi: 10.1002/arch.21992. Epub 2022 Dec 27. Arch Insect Biochem Physiol. 2023. PMID: 36575628 Review.
-
Rice Responses and Resistance to Planthopper-Borne Viruses at Transcriptomic and Proteomic Levels.Curr Issues Mol Biol. 2016;19:43-52. Epub 2015 Sep 11. Curr Issues Mol Biol. 2016. PMID: 26363817 Review.
Cited by
-
A plant virus manipulates the long-winged morph of insect vectors.Proc Natl Acad Sci U S A. 2024 Jan 16;121(3):e2315341121. doi: 10.1073/pnas.2315341121. Epub 2024 Jan 8. Proc Natl Acad Sci U S A. 2024. PMID: 38190519 Free PMC article.
-
Transcript Complexity and New Insights of Restorer Line in CMS-D8 Cotton Through Full-Length Transcriptomic Analysis.Front Plant Sci. 2022 Jun 21;13:930131. doi: 10.3389/fpls.2022.930131. eCollection 2022. Front Plant Sci. 2022. PMID: 35800603 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

