Network Meta-analysis on the Changes of Amyloid Precursor Protein Expression Following SARS-CoV-2 Infection
- PMID: 34757528
- PMCID: PMC8579188
- DOI: 10.1007/s11481-021-10012-9
Network Meta-analysis on the Changes of Amyloid Precursor Protein Expression Following SARS-CoV-2 Infection
Abstract
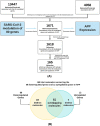
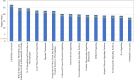
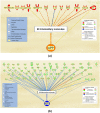
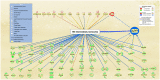
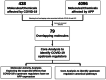
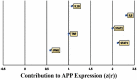
SARS-CoV-2 infection begins with the attachment of its spike (S) protein to angiotensin-converting enzyme-2 (ACE2) followed by complex host immune responses with cardiovascular and neurological implications. Our meta-analyses used QIAGEN Ingenuity Pathway Analysis (IPA) and Knowledge Base (QKB) to investigate how the expression of amyloid precursor protein (APP) was modulated by attachment of SARS-CoV-2 S protein in the brain microvascular endothelial cells (BMVECs) and during COVID-19 in progress. Published 80 host response genes reported to be modulated in BMVECs following SARS-CoV-2 S protein binding were used to identify key canonical pathways and intermediate molecules mediating the regulation of APP production following the attachment of S protein to endothelial cells. This revealed that the attachment of SARS-CoV-2 S protein may inhibit APP expression in the BMVECs. Our results shed light on the molecular mechanisms by which SARS-CoV-2 infection may potentiate the incidence of stroke by inhibiting the production of APP in the BMVECs. We also analyzed molecules associated with COVID-19, which revealed six upstream regulators, TNF, IFNG, STAT1, IL1β, IL6, and STAT3. The upstream regulators mediate the increased production of APP via intermediators, with eleven regulated by all six upstream regulators. These COVID-19 upstream regulators increased APP expression with a statistically significant Z-score of 3.705 (p value = 0.000211). These findings have revealed molecular mechanisms by which COVID-19 disease may lead to long-term neurological manifestations resulting from the elevated APP expression in line with immune response in the host. Altogether, our study revealed two distinct scenarios which may have differential impact on APP expression.
Keywords: Alzheimer’s disease; Blood–brain barrier; Brain microvascular endothelial cells; COVID-19; Neuroinflammation.
© 2021. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Meta-Analysis of APP Expression Modulated by SARS-CoV-2 Infection via the ACE2 Receptor.Int J Mol Sci. 2022 Jan 21;23(3):1182. doi: 10.3390/ijms23031182. Int J Mol Sci. 2022. PMID: 35163117 Free PMC article.
-
SARS-CoV-2 Spike Protein Destabilizes Microvascular Homeostasis.Microbiol Spectr. 2021 Dec 22;9(3):e0073521. doi: 10.1128/Spectrum.00735-21. Epub 2021 Dec 22. Microbiol Spectr. 2021. PMID: 34935423 Free PMC article.
-
Meta-Analysis of the Mechanisms Underlying COVID-19 Modulation of Parkinson's Disease.Int J Mol Sci. 2023 Aug 31;24(17):13554. doi: 10.3390/ijms241713554. Int J Mol Sci. 2023. PMID: 37686360 Free PMC article.
-
The expression of hACE2 receptor protein and its involvement in SARS-CoV-2 entry, pathogenesis, and its application as potential therapeutic target.Tumour Biol. 2021;43(1):177-196. doi: 10.3233/TUB-200084. Tumour Biol. 2021. PMID: 34420993 Review.
-
Does SARS-Cov-2 invade the brain? Translational lessons from animal models.Eur J Neurol. 2020 Sep;27(9):1764-1773. doi: 10.1111/ene.14277. Epub 2020 May 22. Eur J Neurol. 2020. PMID: 32333487 Free PMC article. Review.
Cited by
-
Coupling of Alzheimer's Disease Genetic Risk Factors with Viral Susceptibility and Inflammation.Aging Dis. 2024 Oct 1;15(5):2028-2050. doi: 10.14336/AD.2023.1017. Aging Dis. 2024. PMID: 37962454 Free PMC article. Review.
-
Amyloid precursor protein facilitates SARS-CoV-2 virus entry into cells and enhances amyloid-β-associated pathology in APP/PS1 mouse model of Alzheimer's disease.Transl Psychiatry. 2023 Dec 16;13(1):396. doi: 10.1038/s41398-023-02692-z. Transl Psychiatry. 2023. PMID: 38104129 Free PMC article.
-
The relationship of early- and late-onset Alzheimer's disease genes with COVID-19.J Neural Transm (Vienna). 2022 Jul;129(7):847-859. doi: 10.1007/s00702-022-02499-0. Epub 2022 Apr 16. J Neural Transm (Vienna). 2022. PMID: 35429259 Free PMC article. Review.
-
Neurological, psychological, psychosocial complications of long-COVID and their management.Neurol Sci. 2025 Jan;46(1):1-23. doi: 10.1007/s10072-024-07854-5. Epub 2024 Nov 9. Neurol Sci. 2025. PMID: 39516425 Free PMC article. Review.
-
The Neuroimmune Pharmacology of SARS-CoV-2.J Neuroimmune Pharmacol. 2021 Dec;16(4):699-705. doi: 10.1007/s11481-021-10038-z. Epub 2021 Dec 22. J Neuroimmune Pharmacol. 2021. PMID: 34935110 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

