RENATE: A Pseudo-retrosynthetic Tool for Synthetically Accessible de novo Design
- PMID: 34750989
- PMCID: PMC9285524
- DOI: 10.1002/minf.202100207
RENATE: A Pseudo-retrosynthetic Tool for Synthetically Accessible de novo Design
Abstract
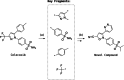
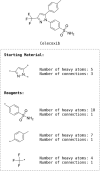
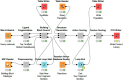
Reaction-based de novo design refers to the generation of synthetically accessible molecules using transformation rules extracted from known reactions in the literature. In this context, we have previously described the extraction of reaction vectors from a reactions database and their coupling with a structure generation algorithm for the generation of novel molecules from a starting material. An issue when designing molecules from a starting material is the combinatorial explosion of possible product molecules that can be generated, especially for multistep syntheses. Here, we present the development of RENATE, a reaction-based de novo design tool, which is based on a pseudo-retrosynthetic fragmentation of a reference ligand and an inside-out approach to de novo design. The reference ligand is fragmented; each fragment is used to search for similar fragments as building blocks; the building blocks are combined into products using reaction vectors; and a synthetic route is suggested for each product molecule. The RENATE methodology is presented followed by a retrospective validation to recreate a set of approved drugs. Results show that RENATE can generate very similar or even identical structures to the corresponding input drugs, hence validating the fragmentation, search, and design heuristics implemented in the tool.
Keywords: de novo drug design; patents; pharmaceuticals; reaction informatics.
© 2021 The Authors. Molecular Informatics published by Wiley-VCH GmbH.
Conflict of interest statement
None declared.
Figures
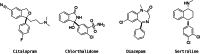
Similar articles
-
NAOMInext - Synthetically feasible fragment growing in a structure-based design context.Eur J Med Chem. 2019 Feb 1;163:747-762. doi: 10.1016/j.ejmech.2018.11.075. Epub 2018 Dec 6. Eur J Med Chem. 2019. PMID: 30576905
-
Enhancing reaction-based de novo design using a multi-label reaction class recommender.J Comput Aided Mol Des. 2020 Jul;34(7):783-803. doi: 10.1007/s10822-020-00300-6. Epub 2020 Feb 28. J Comput Aided Mol Des. 2020. PMID: 32112286 Free PMC article.
-
Flux (1): a virtual synthesis scheme for fragment-based de novo design.J Chem Inf Model. 2006 Mar-Apr;46(2):699-707. doi: 10.1021/ci0503560. J Chem Inf Model. 2006. PMID: 16563000
-
Recent Advances in Automated Structure-Based De Novo Drug Design.J Chem Inf Model. 2024 Mar 25;64(6):1794-1805. doi: 10.1021/acs.jcim.4c00247. Epub 2024 Mar 14. J Chem Inf Model. 2024. PMID: 38485516 Free PMC article. Review.
-
Concepts and applications of "natural computing" techniques in de novo drug and peptide design.Curr Pharm Des. 2010 May;16(15):1656-65. doi: 10.2174/138161210791164009. Curr Pharm Des. 2010. PMID: 20222857 Review.
Cited by
-
MegaSyn: Integrating Generative Molecular Design, Automated Analog Designer, and Synthetic Viability Prediction.ACS Omega. 2022 May 27;7(22):18699-18713. doi: 10.1021/acsomega.2c01404. eCollection 2022 Jun 7. ACS Omega. 2022. PMID: 35694522 Free PMC article.
-
Synthetically accessible de novo design using reaction vectors: Application to PARP1 inhibitors.Mol Inform. 2024 Apr;43(4):e202300183. doi: 10.1002/minf.202300183. Epub 2024 Feb 6. Mol Inform. 2024. PMID: 38258328 Free PMC article.
-
A molecule perturbation software library and its application to study the effects of molecular design constraints.J Cheminform. 2023 Sep 26;15(1):89. doi: 10.1186/s13321-023-00761-5. J Cheminform. 2023. PMID: 37752561 Free PMC article.
References
-
- Hartenfeller M., Schneider G., Hartenfeller M., Proschak E., De novo Drug Design. In: Bajorath J (ed) Lead Generation Approaches in Drug Discovery. John Wiley & Sons, Inc., Hoboken, NJ, USA, 2010, pp 165–185.
-
- Schneider P., Schneider G., J. Med. Chem. 2016, 59, 4077–4086. doi: 10.1021/acs.jmedchem.5b01849. - PubMed
-
- Gasteiger J., J. Comput.-Aided Mol. Des. 2007, 21, 307–309. doi: 10.1007/s10822-007-9115–1. - PubMed
-
- Vinkers H. M., de Jonge M. R., Daeyaert F. F. D., et al., J. Med. Chem. 2003, 46, 2765–2773. doi: 10.1021/jm030809x. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

