Site-specific SUMOylation of viral polymerase processivity factor: a way of localizingtoND10 subnuclear domains for restricted and self-controlled reproduction of herpesvirus
- PMID: 34747321
- PMCID: PMC8923073
- DOI: 10.1080/21505594.2021.2000689
Site-specific SUMOylation of viral polymerase processivity factor: a way of localizingtoND10 subnuclear domains for restricted and self-controlled reproduction of herpesvirus
Abstract
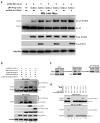
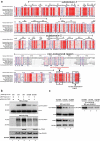
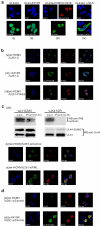
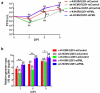
Lytic replication of human cytomegalovirus (HCMV), a member of β-herpesvirus, is a highly complicated and organized process that requires its DNA polymerase processivity factor, UL44, the first-reported HCMV replication protein subjected to SUMO post-translational modification (PTM). SUMOylation plays a pleiotropic role in protein functions of host cells and infecting viruses. Particularly, formation of herpesviral replication compartments (RCs) upon infection is induced in proximity to ND10 subnuclear domains, the host cell's intrinsic antiviral immune devices and hot SUMOylation spots, relying just on SUMOylation of their protein components to become mature and functional in restriction of the viral replication. In this study, to unveil the exact role of SUMO PTM on UL44 involved in HCMV replication, we screened and identified PIAS3, an annotated E3 SUMO ligase, as a novel UL44-interacting protein engaged in cellular SUMOylation pathway. Co-existence of PIAS3 could enhance the UBC9-based SUMO modification of UL44 specifically at its conserved 410lysine residue lying within the single canonical ψKxE SUMO Conjugation Motif (SCM). Intriguingly, we found this SCM-specific SUMOylation contributes to UL44 co-localization and interaction with subnuclear ND10 domains during infection, which in turn exerts an inhibitory effect on HCMV replication and growth. Together, these results highlight the importance of SUMOylation in regulating viral protein subnuclear localization, representing a novel way of utilizing ND10-based restriction to achieve the self-controlled slower replication and reproduction of herpesviruses.
Keywords: E3 SUMO ligase; Human cytomegalovirus (HCMV); PIAS3; SUMO conjugation motif (SCM); SUMOylation; UL44; subnuclear localization, ND10; viral polymerase processivity factor.
Conflict of interest statement
The authors have declared no competing financial interests exist.
Figures


Similar articles
-
Sumoylation of the Carboxy-Terminal of Human Cytomegalovirus DNA Polymerase Processivity Factor UL44 Attenuates Viral DNA Replication.Front Microbiol. 2021 Apr 21;12:652719. doi: 10.3389/fmicb.2021.652719. eCollection 2021. Front Microbiol. 2021. PMID: 33967989 Free PMC article.
-
The ND10 Component Promyelocytic Leukemia Protein Acts as an E3 Ligase for SUMOylation of the Major Immediate Early Protein IE1 of Human Cytomegalovirus.J Virol. 2017 Apr 28;91(10):e02335-16. doi: 10.1128/JVI.02335-16. Print 2017 May 15. J Virol. 2017. PMID: 28250117 Free PMC article.
-
The human cytomegalovirus DNA polymerase processivity factor UL44 is modified by SUMO in a DNA-dependent manner.PLoS One. 2012;7(11):e49630. doi: 10.1371/journal.pone.0049630. Epub 2012 Nov 15. PLoS One. 2012. PMID: 23166733 Free PMC article.
-
Viruses, SUMO, and immunity: the interplay between viruses and the host SUMOylation system.J Neurovirol. 2021 Aug;27(4):531-541. doi: 10.1007/s13365-021-00995-9. Epub 2021 Aug 3. J Neurovirol. 2021. PMID: 34342851 Free PMC article. Review.
-
SUMOylation in Viral Replication and Antiviral Defense.Adv Sci (Weinh). 2022 Mar;9(7):e2104126. doi: 10.1002/advs.202104126. Epub 2022 Jan 21. Adv Sci (Weinh). 2022. PMID: 35060688 Free PMC article. Review.
Cited by
-
The SUMOylation of Human Cytomegalovirus Capsid Assembly Protein Precursor (UL80.5) Affects Its Interaction with Major Capsid Protein (UL86) and Viral Replication.Viruses. 2023 Apr 7;15(4):931. doi: 10.3390/v15040931. Viruses. 2023. PMID: 37112911 Free PMC article.
-
Enzymatic independent role of sphingosine kinase 2 in regulating the expression of type I interferon during influenza A virus infection.PLoS Pathog. 2022 Sep 7;18(9):e1010794. doi: 10.1371/journal.ppat.1010794. eCollection 2022 Sep. PLoS Pathog. 2022. PMID: 36070294 Free PMC article.
References
-
- Mocarski ES, Shenk T, Griffiths PD, et al. Cytomegaloviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 6th ed. Vol. 2. Lippincott Williams and Williams,Philadelphia, PA, USA; 2013, p. 1960–2014.
-
- Weiland KL, Oien NL, Homa F, et al. Functional analysis of human cytomegalovirus polymerase accessory protein. Virus Res. 1994;34(3):191–206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
