Molecular conformations and dynamics in the extracellular matrix of mammalian structural tissues: Solid-state NMR spectroscopy approaches
- PMID: 34746737
- PMCID: PMC8551230
- DOI: 10.1016/j.mbplus.2021.100086
Molecular conformations and dynamics in the extracellular matrix of mammalian structural tissues: Solid-state NMR spectroscopy approaches
Abstract
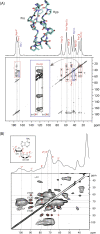
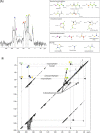
Solid-state NMR spectroscopy has played an important role in multidisciplinary studies of the extracellular matrix. Here we review how solid-state NMR has been used to probe collagen molecular conformations, dynamics, post-translational modifications and non-enzymatic chemical changes, and in calcified tissues, the molecular structure of bone mineral and its interface with collagen. We conclude that NMR spectroscopy can deliver vital information that in combination with data from structural imaging techniques, can result in significant new insight into how the extracellular matrix plays its multiple roles.
Keywords: Biomineralization; Bone mineral; Collagen; Extracellular matrix structure; Multidimensional solid-state NMR spectroscopy; Proline conformation.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Collagen Structure-Function Relationships from Solid-State NMR Spectroscopy.Acc Chem Res. 2018 Jul 17;51(7):1621-1629. doi: 10.1021/acs.accounts.8b00092. Epub 2018 Jun 22. Acc Chem Res. 2018. PMID: 29931970 Review.
-
The contribution of solid-state NMR spectroscopy to understanding biomineralization: atomic and molecular structure of bone.J Magn Reson. 2015 Apr;253:98-110. doi: 10.1016/j.jmr.2014.12.011. J Magn Reson. 2015. PMID: 25797009
-
31P and 13C solid-state NMR spectroscopy to study collagen synthesis and biomineralization in polymer-based bone implants.NMR Biomed. 2012 Mar;25(3):464-75. doi: 10.1002/nbm.1649. Epub 2011 Jan 12. NMR Biomed. 2012. PMID: 22351643
-
Cell adhesion promoting peptide GVKGDKGNPGWPGAP from the collagen type IV triple helix: cis/trans proline-induced multiple 1H NMR conformations and evidence for a KG/PG multiple turn repeat motif in the all-trans proline state.Biochemistry. 1991 Aug 20;30(33):8251-67. doi: 10.1021/bi00247a022. Biochemistry. 1991. PMID: 1868097
-
Proton-Based Ultrafast Magic Angle Spinning Solid-State NMR Spectroscopy.Acc Chem Res. 2017 Apr 18;50(4):1105-1113. doi: 10.1021/acs.accounts.7b00082. Epub 2017 Mar 29. Acc Chem Res. 2017. PMID: 28353338 Free PMC article. Review.
Cited by
-
Age-related changes in cationic compositions of human cranial base bone apatite measured by X-ray energy dispersive spectroscopy (EDS) coupled with scanning electron microscope (SEM).Biometals. 2022 Oct;35(5):1077-1094. doi: 10.1007/s10534-022-00425-1. Epub 2022 Aug 4. Biometals. 2022. PMID: 35922585
-
Resolving Atomic-Level Dynamics and Interactions of High-Molecular-Weight Hyaluronic Acid by Multidimensional Solid-State NMR.ACS Appl Mater Interfaces. 2024 Aug 21;16(33):43317-43328. doi: 10.1021/acsami.4c08428. Epub 2024 Aug 9. ACS Appl Mater Interfaces. 2024. PMID: 39121380 Free PMC article.
-
Nuclear magnetic resonance spectroscopy to quantify major extracellular matrix components in fibro-calcific aortic valve disease.Sci Rep. 2023 Nov 1;13(1):18823. doi: 10.1038/s41598-023-46143-7. Sci Rep. 2023. PMID: 37914797 Free PMC article.
-
Unveiling Charge-Pair Salt-Bridge Interaction Between GAGs and Collagen Protein in Cartilage: Atomic Evidence from DNP-Enhanced ssNMR at Natural Isotopic Abundance.J Am Chem Soc. 2024 Aug 28;146(34):23663-23668. doi: 10.1021/jacs.4c05539. Epub 2024 Jul 9. J Am Chem Soc. 2024. PMID: 38980938
References
-
- Wishart D.S., Sykes B.D., Richards F.M. The Chemical shift index. A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31(1992):1647–1651. - PubMed
-
- Böckmann Anja. Structural and dynamic studies of proteins by high-resolution solid-state NMR. C. R. Chim. Mar 2006;9(3–4):381–392.
Publication types
LinkOut - more resources
Full Text Sources

