Dishevelled-1 DIX and PDZ domain lysine residues regulate oncogenic Wnt signaling
- PMID: 34733415
- PMCID: PMC8555683
- DOI: 10.18632/oncotarget.28089
Dishevelled-1 DIX and PDZ domain lysine residues regulate oncogenic Wnt signaling
Abstract
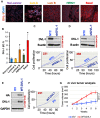
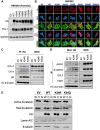
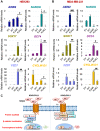
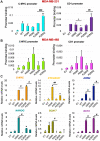
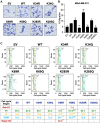
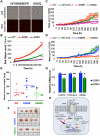
DVL proteins are central mediators of the Wnt pathway and relay complex input signals into different branches of the Wnt signaling network. However, molecular mechanism(s) that regulate DVL-mediated relay of Wnt signals still remains unclear. Here, for the first time, we elucidate the functional significance of three DVL-1 lysines (K/Lys) which are subject to post-translational acetylation. We demonstrate that K34 Lys residue in the DIX domain regulates subcellular localization of β-catenin, thereby influencing downstream Wnt target gene expression. Additionally, we show that K69 (DIX domain) and K285 (PDZ domain) regulate binding of DVL-1 to Wnt target gene promoters and modulate expression of Wnt target genes including CMYC, OCT4, NANOG, and CCND1, in cell line models and xenograft tumors. Finally, we report that conserved DVL-1 lysines modulate various oncogenic functions such as cell migration, proliferation, cell-cycle progression, 3D-spheroid formation and in-vivo tumor growth in breast cancer models. Collectively, these findings highlight the importance of DVL-1 domain-specific lysines which were recently shown to be acetylated and characterize their influence on Wnt signaling. These site-specific modifications may be subject to regulation by therapeutics already in clinical use (lysine deacetylase inhibitors such as Panobinostat and Vorinostat) or may possibly have prognostic utility in translational efforts that seek to modulate dysfunctional Wnt signaling.
Keywords: chromatin immunoprecipitation (ChIP); dishevelled (DVL); gene expression; lysine residue; post-translational modification.
Copyright: © 2021 Sharma et al.
Conflict of interest statement
CONFLICTS OF INTEREST Authors have no conflicts of interest to declare.
Figures
Similar articles
-
Acetylation of conserved DVL-1 lysines regulates its nuclear translocation and binding to gene promoters in triple-negative breast cancer.Sci Rep. 2019 Nov 7;9(1):16257. doi: 10.1038/s41598-019-52723-3. Sci Rep. 2019. PMID: 31700102 Free PMC article.
-
Autoinhibition of Dishevelled protein regulated by its extreme C terminus plays a distinct role in Wnt/β-catenin and Wnt/planar cell polarity (PCP) signaling pathways.J Biol Chem. 2017 Apr 7;292(14):5898-5908. doi: 10.1074/jbc.M116.772509. Epub 2017 Feb 21. J Biol Chem. 2017. PMID: 28223363 Free PMC article.
-
Can We Pharmacologically Target Dishevelled: The Key Signal Transducer in the Wnt Pathways?Handb Exp Pharmacol. 2021;269:117-135. doi: 10.1007/164_2021_527. Handb Exp Pharmacol. 2021. PMID: 34382124
-
Regulation of Dishevelled protein activity and stability by post-translational modifications and autophagy.Trends Biochem Sci. 2021 Dec;46(12):1003-1016. doi: 10.1016/j.tibs.2021.07.008. Epub 2021 Aug 23. Trends Biochem Sci. 2021. PMID: 34433516 Review.
-
Dishevelled: An emerging therapeutic oncogene in human cancers.Pathol Res Pract. 2023 Oct;250:154793. doi: 10.1016/j.prp.2023.154793. Epub 2023 Sep 4. Pathol Res Pract. 2023. PMID: 37683388 Review.
Cited by
-
Dishevelled 2 regulates cancer cell proliferation and T cell mediated immunity in HER2-positive breast cancer.BMC Cancer. 2023 Feb 21;23(1):172. doi: 10.1186/s12885-023-10647-2. BMC Cancer. 2023. PMID: 36809986 Free PMC article.
-
Dishevelled localization and function are differentially regulated by structurally distinct sterols.bioRxiv [Preprint]. 2024 May 15:2024.05.14.593701. doi: 10.1101/2024.05.14.593701. bioRxiv. 2024. PMID: 38798572 Free PMC article. Preprint.
-
Dishevelled phase separation promotes Wnt signalosome assembly and destruction complex disassembly.J Cell Biol. 2022 Dec 5;221(12):e202205069. doi: 10.1083/jcb.202205069. Epub 2022 Nov 7. J Cell Biol. 2022. PMID: 36342472 Free PMC article.
-
Yin Yang 1 promotes the neuroendocrine differentiation of prostate cancer cells via the non-canonical WNT pathway (FYN/STAT3).Clin Transl Med. 2023 Oct;13(10):e1422. doi: 10.1002/ctm2.1422. Clin Transl Med. 2023. PMID: 37771187 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

