PANTHER: Making genome-scale phylogenetics accessible to all
- PMID: 34717010
- PMCID: PMC8740835
- DOI: 10.1002/pro.4218
PANTHER: Making genome-scale phylogenetics accessible to all
Abstract
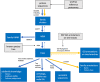
Phylogenetics is a powerful tool for analyzing protein sequences, by inferring their evolutionary relationships to other proteins. However, phylogenetics analyses can be challenging: they are computationally expensive and must be performed carefully in order to avoid systematic errors and artifacts. Protein Analysis THrough Evolutionary Relationships (PANTHER; http://pantherdb.org) is a publicly available, user-focused knowledgebase that stores the results of an extensive phylogenetic reconstruction pipeline that includes computational and manual processes and quality control steps. First, fully reconciled phylogenetic trees (including ancestral protein sequences) are reconstructed for a set of "reference" protein sequences obtained from fully sequenced genomes of organisms across the tree of life. Second, the resulting phylogenetic trees are manually reviewed and annotated with function evolution events: inferred gains and losses of protein function along branches of the phylogenetic tree. Here, we describe in detail the current contents of PANTHER, how those contents are generated, and how they can be used in a variety of applications. The PANTHER knowledgebase can be downloaded or accessed via an extensive API. In addition, PANTHER provides software tools to facilitate the application of the knowledgebase to common protein sequence analysis tasks: exploring an annotated genome by gene function; performing "enrichment analysis" of lists of genes; annotating a single sequence or large batch of sequences by homology; and assessing the likelihood that a genetic variant at a particular site in a protein will have deleterious effects.
Keywords: gene ontology; genome analysis; hidden Markov model; molecular evolution; omics data analysis; phylogenetic tree; protein function annotation; protein function evolution.
© 2021 The Protein Society.
Figures

Similar articles
-
PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium.Nucleic Acids Res. 2010 Jan;38(Database issue):D204-10. doi: 10.1093/nar/gkp1019. Epub 2009 Dec 16. Nucleic Acids Res. 2010. PMID: 20015972 Free PMC article.
-
PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API.Nucleic Acids Res. 2021 Jan 8;49(D1):D394-D403. doi: 10.1093/nar/gkaa1106. Nucleic Acids Res. 2021. PMID: 33290554 Free PMC article.
-
PANTHER version 10: expanded protein families and functions, and analysis tools.Nucleic Acids Res. 2016 Jan 4;44(D1):D336-42. doi: 10.1093/nar/gkv1194. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26578592 Free PMC article.
-
Inferring trees.Methods Mol Biol. 2008;452:287-309. doi: 10.1007/978-1-60327-159-2_14. Methods Mol Biol. 2008. PMID: 18566770 Review.
-
Phylogenetic analysis of general bacterial porins: a phylogenomic case study.J Mol Microbiol Biotechnol. 2006;11(6):291-301. doi: 10.1159/000095631. J Mol Microbiol Biotechnol. 2006. PMID: 17114893 Review.
Cited by
-
Combined Effects of Polystyrene Nanosphere and Homosolate Exposures on Estrogenic End Points in MCF-7 Cells and Zebrafish.Environ Health Perspect. 2024 Feb;132(2):27011. doi: 10.1289/EHP13696. Epub 2024 Feb 21. Environ Health Perspect. 2024. PMID: 38381479 Free PMC article.
-
Spatial multi-omics in whole skeletal muscle reveals complex tissue architecture.Commun Biol. 2024 Oct 5;7(1):1272. doi: 10.1038/s42003-024-06949-1. Commun Biol. 2024. PMID: 39369093 Free PMC article.
-
Genome-wide binding analysis unveils critical implication of B-Myb-mediated transactivation in cancers.Int J Biol Sci. 2024 Sep 3;20(12):4691-4712. doi: 10.7150/ijbs.92607. eCollection 2024. Int J Biol Sci. 2024. PMID: 39309447 Free PMC article.
-
Large-scale genome-wide association study to identify causal relationships and potential mediators between education and autoimmune diseases.Front Immunol. 2023 Dec 7;14:1249017. doi: 10.3389/fimmu.2023.1249017. eCollection 2023. Front Immunol. 2023. PMID: 38146362 Free PMC article.
-
Profiling Tel1 Signaling Reveals a Non-Canonical Motif Targeting DNA Repair and Telomere Control Machineries.bioRxiv [Preprint]. 2024 Jul 5:2024.07.03.601872. doi: 10.1101/2024.07.03.601872. bioRxiv. 2024. Update in: J Biol Chem. 2025 Jan 16:108194. doi: 10.1016/j.jbc.2025.108194 PMID: 39005478 Free PMC article. Updated. Preprint.
References
-
- Legried B, Molloy EK, Warnow T, Roch S. Polynomial‐time statistical estimation of species trees under gene duplication and loss. J Comput Biol. 2021;28:452–468. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

