An Assessment of Serological Assays for SARS-CoV-2 as Surrogates for Authentic Virus Neutralization
- PMID: 34704832
- PMCID: PMC8549747
- DOI: 10.1128/Spectrum.01059-21
An Assessment of Serological Assays for SARS-CoV-2 as Surrogates for Authentic Virus Neutralization
Abstract
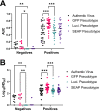
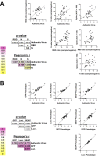
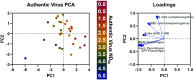
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in late 2019 and has since caused a global pandemic resulting in millions of cases and deaths. Diagnostic tools and serological assays are critical for controlling the outbreak, especially assays designed to quantitate neutralizing antibody levels, considered the best correlate of protection. As vaccines become increasingly available, it is important to identify reliable methods for measuring neutralizing antibody responses that correlate with authentic virus neutralization but can be performed outside biosafety level 3 (BSL3) laboratories. While many neutralizing assays using pseudotyped virus have been developed, there have been few studies comparing the different assays to each other as surrogates for authentic virus neutralization. Here, we characterized three enzyme-linked immunosorbent assays (ELISAs) and three pseudotyped vesicular stomatitis virus (VSV) neutralization assays and assessed their concordance with authentic virus neutralization. The most accurate assays for predicting authentic virus neutralization were luciferase- and secreted embryonic alkaline phosphatase (SEAP)-expressing pseudotyped virus neutralizations, followed by green fluorescent protein (GFP)-expressing pseudotyped virus neutralization, and then the ELISAs. IMPORTANCE The ongoing COVID-19 pandemic is caused by infection with severe acute respiratory syndrome virus 2 (SARS-CoV-2). Prior infection or vaccination can be detected by the presence of antibodies in the blood. Antibodies in the blood are also considered to be protective against future infections from the same virus. The "gold standard" assay for detecting protective antibodies against SARS-CoV-2 is neutralization of authentic SARS-CoV-2 virus. However, this assay can only be performed under highly restrictive biocontainment conditions. We therefore characterized six antibody-detecting assays for their correlation with authentic virus neutralization. The significance of our research is in outlining the advantages and disadvantages of the different assays and identifying the optimal surrogate assay for authentic virus neutralization. This will allow for more accurate assessments of protective immunity against SARS-CoV-2 following infection and vaccination.
Keywords: SARS-CoV-2; immunoassays; neutralizing antibodies.
Figures


Similar articles
-
Evaluation of SARS-CoV-2 neutralizing antibodies using a vesicular stomatitis virus possessing SARS-CoV-2 spike protein.Virol J. 2021 Jan 12;18(1):16. doi: 10.1186/s12985-021-01490-7. Virol J. 2021. PMID: 33435994 Free PMC article.
-
Evaluation of Three Quantitative Anti-SARS-CoV-2 Antibody Immunoassays.Microbiol Spectr. 2021 Dec 22;9(3):e0137621. doi: 10.1128/spectrum.01376-21. Epub 2021 Dec 22. Microbiol Spectr. 2021. PMID: 34937195 Free PMC article.
-
Evaluating Humoral Immunity against SARS-CoV-2: Validation of a Plaque-Reduction Neutralization Test and a Multilaboratory Comparison of Conventional and Surrogate Neutralization Assays.Microbiol Spectr. 2021 Dec 22;9(3):e0088621. doi: 10.1128/Spectrum.00886-21. Epub 2021 Nov 17. Microbiol Spectr. 2021. PMID: 34787495 Free PMC article.
-
Recent Developments in SARS-CoV-2 Neutralizing Antibody Detection Methods.Curr Med Sci. 2021 Dec;41(6):1052-1064. doi: 10.1007/s11596-021-2470-7. Epub 2021 Dec 21. Curr Med Sci. 2021. PMID: 34935114 Free PMC article. Review.
-
Overview of Neutralization Assays and International Standard for Detecting SARS-CoV-2 Neutralizing Antibody.Viruses. 2022 Jul 18;14(7):1560. doi: 10.3390/v14071560. Viruses. 2022. PMID: 35891540 Free PMC article. Review.
Cited by
-
Development and Evaluation of a Monoclonal Antibody-Based Blocking Enzyme-Linked Immunosorbent Assay for the Detection of Antibodies against Novel Duck Reovirus in Waterfowl Species.Microbiol Spectr. 2022 Dec 21;10(6):e0258122. doi: 10.1128/spectrum.02581-22. Epub 2022 Nov 29. Microbiol Spectr. 2022. PMID: 36445088 Free PMC article.
-
Host Predictors of Broadly Cross-Reactive Antibodies Against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Variants of Concern Differ Between Infection and Vaccination.Clin Infect Dis. 2022 Aug 24;75(1):e705-e714. doi: 10.1093/cid/ciab996. Clin Infect Dis. 2022. PMID: 34891165 Free PMC article.
-
Correlation between pseudotyped virus and authentic virus neutralisation assays, a systematic review and meta-analysis of the literature.Front Immunol. 2023 Sep 18;14:1184362. doi: 10.3389/fimmu.2023.1184362. eCollection 2023. Front Immunol. 2023. PMID: 37790941 Free PMC article.
-
What can neutralizing antibodies tell us about the quality of immunity in COVID-19 convalescents and vaccinees?Hum Vaccin Immunother. 2023 Dec 15;19(3):2270310. doi: 10.1080/21645515.2023.2270310. Epub 2023 Oct 31. Hum Vaccin Immunother. 2023. PMID: 37905722 Free PMC article.
-
Utility of nasal swabs for assessing mucosal immune responses towards SARS-CoV-2.Sci Rep. 2023 Oct 19;13(1):17820. doi: 10.1038/s41598-023-44989-5. Sci Rep. 2023. PMID: 37857783 Free PMC article.
References
-
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette J-L, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. doi:10.2807/1560-7917.ES.2020.25.3.2000045. - DOI - PMC - PubMed
-
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi:10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
-
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi:10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Grzelak L, Temmam S, Planchais C, Demeret C, Tondeur L, Huon C, Guivel-Benhassine F, Staropoli I, Chazal M, Dufloo J, Planas D, Buchrieser J, Rajah MM, Robinot R, Porrot F, Albert M, Chen K-Y, Crescenzo-Chaigne B, Donati F, Anna F, Souque P, Gransagne M, Bellalou J, Nowakowski M, Backovic M, Bouadma L, Le Fevre L, Le Hingrat Q, Descamps D, Pourbaix A, Laouénan C, Ghosn J, Yazdanpanah Y, Besombes C, Jolly N, Pellerin-Fernandes S, Cheny O, Ungeheuer M-N, Mellon G, Morel P, Rolland S, Rey FA, Behillil S, Enouf V, Lemaitre A, Créach M-A, Petres S, Escriou N, Charneau P, Fontanet A, et al. . 2020. A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations. Sci Transl Med 12:eabc3103. doi:10.1126/scitranslmed.abc3103. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

